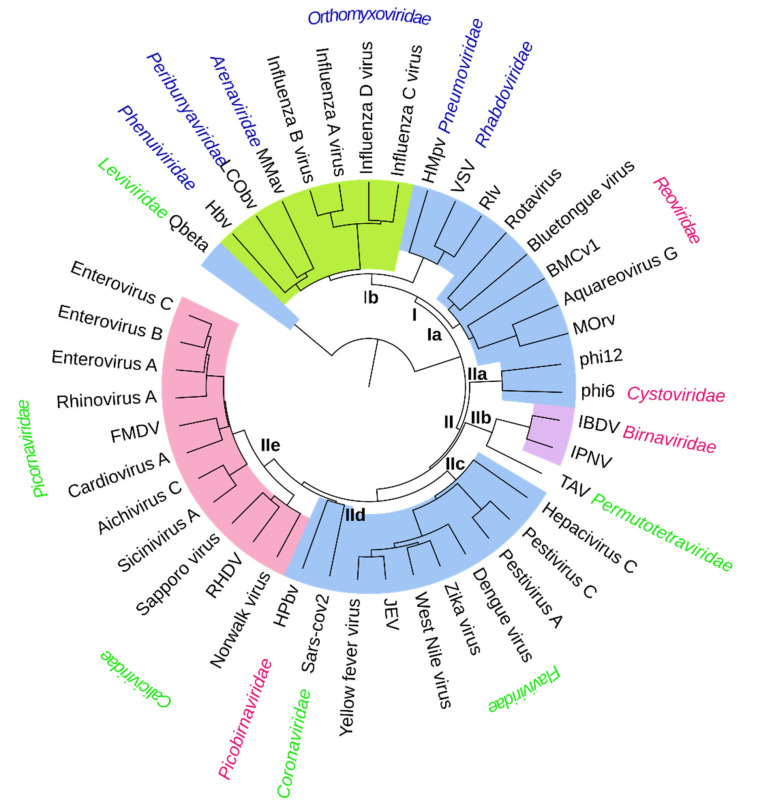
Figure 2.
A structure-based phylogenetic tree for the polymerase subunits of RNA viruses deduced based on the structural alignment of the common core. The name of the virus (black) and the viral family are indicated at the tips of the branches. The coloring of the family name reflects the nature of the viral genome: green, positive-strand RNA; blue, negative-strand RNA; and red, double-stranded RNA. The polymerases using solely primer-independent initiation are indicated with blue background, polymerases using VPg-protein primer with pink background, polymerases applying self-priming with purple background, and polymerases using RNA-priming on (+)strand synthesis with green background. Background is white if the initiation mechanism is unclear. Moreover, in this case, there is biochemical and structural evidence that suggests a primer-independent initiation mechanism [43]. The applied abbreviations are: BMCv1: Bombyx Mori cypovirus 1, FMDV: foot-and-mouth disease virus, Hbv: Huaiyangshan banyangvirus, HPbv: human picobirnavirus, HMpv: human metapneumovirus, IBDV: infectious bursal disease virus, IPNV: infectious pancreatic necrosis virus, JEV: Japanese encephalitis virus, LCObv: La Crosse orthobunyavirus, MMav: Machupo mammarenavirus, MOrv: mammalian orthoreovirus, phi6: Pseudomonas phage phi6, phi12: Pseudomonas phage phi12, Qbeta: Escherichia phage Qbeta, RHDV: rabbit haemorrhagic disease virus, Rlv: rabies lyssavirus, SARS-CoV-2: severe acute respiratory syndrome coronavirus 2, TAV: thosean asigna virus, VSV: vesicular stomatitis Indian virus.