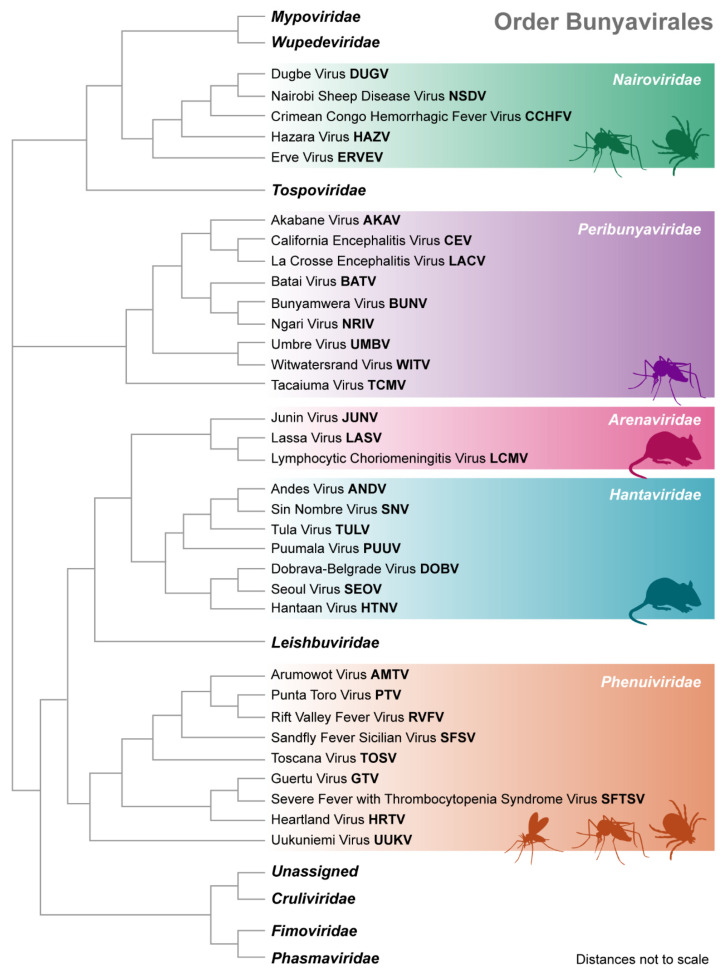
Figure 1.
Bunyavirales Phylogenetic Tree. Order Bunyavirales phylogenetic tree based on nucleoprotein amino acid sequences of the 13 families. Families for which a specific virus is not listed are arranged via analysis of type species as listed in Table 1. Specific vector species are depicted for Nairoviridae (mosquito, tick), Peribunyaviridae (mosquito), Arenaviridae (rodent), Hantaviridae (rodent), and Phenuiviridae (sandfly, mosquito, tick). While these families may contain viruses transmitted through other vectors, these illustrations represent those of the viruses listed and discussed in this review. The phylogenetic tree was constructed using S segment nucleoprotein amino acid sequences from Genbank and was assembled using Geneious Prime tree builder global alignment with free end gaps, cost matrix PAM250, genetic distance model Jukes-Cantor, tree build method neighbor-joining, and no outgroup. The resulting tree was transformed so that branches are of equal length. Tree segments are not to scale. Genbank accession numbers for sequences used: Mypoviridae (NC_033760.1), Wupedeviridae (NC_043501.1), Nairoviridae (MH483984.1, FJ422213.2, MH791451.1, NC_038711.1, JF911699.1), Tospoviridae (MN861976.1), Peribunyaviridae (MH484290.1, MT276603.1, MH830340.1, MT022508.1, KM507341.1, MK896460.1, MK330166.1, NC_043673.1, LC552050.1), Arenaviridae (MG554174.1, MG189700.1, MT861994.1), Hantaviridae (MN258229.1, MT514275.1, MN832781.1, MN657233.1, MK360773.1, MT012546.1, KT885046.1), Leishbuviridae (KX280017.1), Phenuiviridae (DQ380149.1, NC_018137.1, NC_024496.1, EF201835.1, MT032306.1, KM114248.1, NC_043610.1, HM566145.1, EF201822.1), Cruliviridae (NC_032145.1), Fimoviridae (LR536377.1), Phasmaviridae (NC_043032.1), and Unassigned (MG764564.1).