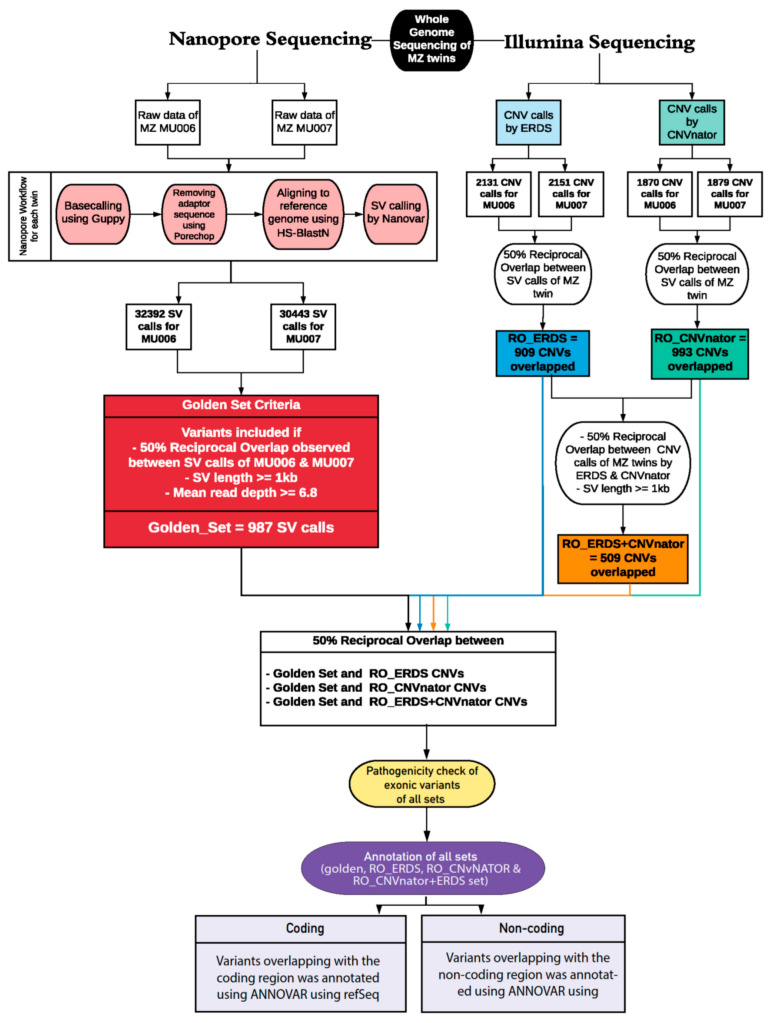
Figure 1.
Overview of the Structural Variant Calling workflow for whole-genome sequencing data performed by two technologies—long-read sequencing by nanopore sequencing and short-read sequencing by llumina sequencing—for detection of structural variants in monozygotic twins (MU006 and MU007). For the first analysis, the raw fast5 files acquired from Oxford Nanopore Technologies (ONT) sequencer were processed using the following steps for each monozygotic twin: base calling done by Guppy, filtering by Porechop, mapping to hg38 reference genome using HS-BlastN, and Nanovar was used for calling structural variant. For the second analysis, the CNV calls from Illumina sequencing were established using two callers: CNVnator and ERDS caller. To determine identical variants between the twins for each caller—Nanovar, CNVnator, and ERDS—we performed a 50% reciprocal overlap analysis on their VCF outputs and filtered out variants below 1 kb to formulate the ‘golden set’, RO_CNVnator set, and RO_ERDS sets, respectively. To investigate identical variants between the LRS and SRS platforms, reciprocal over-lap analysis was done on the ‘golden set’, RO_CNVnator, and RO_ERDS sets. All sets were investigated for pathogenicity and annotated using Annovar to determine the coding and non-coding genomic elements overlapping with the examined variants.