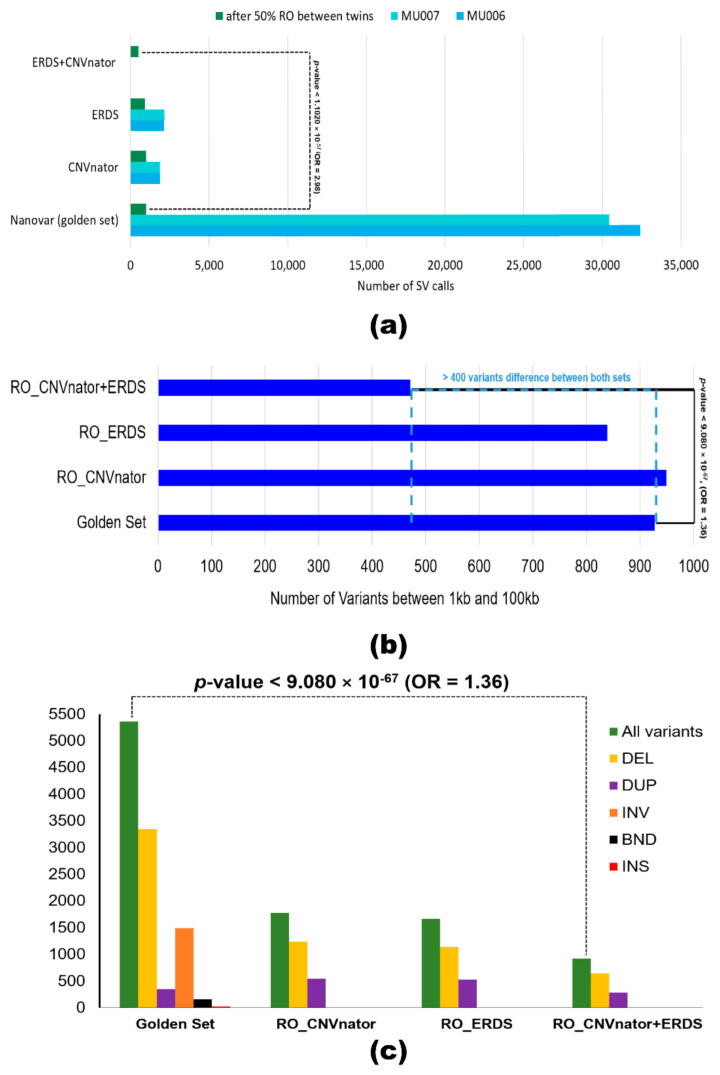
Figure 4.
Comparison between variant sets formulated by polishing LRS calls (‘golden set’) and SRS calls (RO_CNVnator, RO_ERDS, and RO_CNVnator + ERDS sets). (a) Bar graph representing the number of variants called for each of the monozygotic twins and after their reciprocal overlap analysis by callers—Nanovar (in green), ERDS (light blue), and CNVnator (dark blue). (b) Assessment of the number of structural variants present within the 1 to 100 kb size range for each set is represented in a bar graph. (c) Comparison between the number of genes overlapping with variants called by the different sets of LRS and SRS callers and a breakdown of the gene distribution per variant type—deletions (in yellow), duplications (in purple), inversion (in orange), breakends (in black) and insertions (in red). The p-value and odds ratio show the association between long-read caller NanoVar and its ability to call a number of variants (a) within the 1–100 kb size range, (b) affecting the number of genes, and (c) when compared to illumina consensus set.