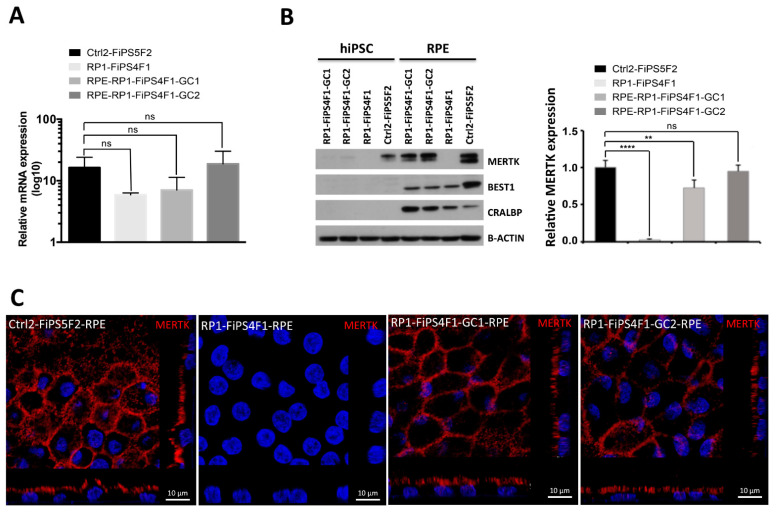
Figure 3.
Recovery of Mer tyrosine kinase receptor (MERTK) expression in vitro. (A) qRT-PCR analysis of mRNA expression for MERTK. Data represent fold-change in mRNA expression of four hiPSC-RPE lines relative to their hiPSC. Each bar represents the average ±SEM of at least three independent biological replicates. The differences in mRNA expression between the control hiPSC-RPE with the uncorrected and corrected RP-hiPSC-RPE were tested for significance. (B) Western blot analysis of RPE-specific marker protein expression in hiPSC-RPE. The expression of CRALBP and BEST1 is detected in hiPSC-RPE, but not in the respective parental hiPSCs. MERTK expression is detected in corrected and control hiPSC-RPE but not in uncorrected RP-hiPSC-RPE. β-ACTIN used as loading control. The graph shows the relative quantification of MERTK expression representative of three different experiments of western blot. The differences in protein expression between the control hiPSC-RPE with the uncorrected and corrected RP-hiPSC-RPE were tested for significance (** p ≤ 0.01, **** p ≤ 0.0001) (C) MERTK expression in hiPSC-RPE. Confocal z-stacks micrographs show apical MERTK distribution in gene-corrected RP-hiPSC-RPE (RP1-FiPS4F1-GC1 and RP1-FiPS4F1-GC2) and healthy control hiPSC-RPE (Ctrl2-FiPS5F2). We failed to detect MERTK expression in uncorrected RP-hiPSC-RPE (RP1-FiPS4F1).