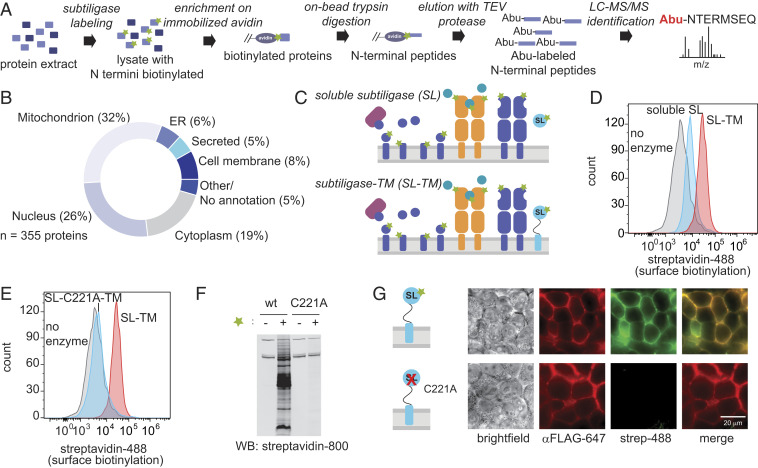
Fig. 1.
Restricting subtiligase activity to the cell surface. (A) Workflow for subtiligase N terminomics in cell lysate. Biotinylated peptide ester is represented by a green star. (B) Subcellular locations of N termini identified in a subtiligase N terminomics experiment performed using soluble subtiligase in HEK293T cell lysate. (C) Approaches for labeling cell surface N termini. (Top) Addition of soluble subtiligase to live cells. (Bottom) Expression of subtiligase fused to the PDGFRβ chain (subtiligase-TM) in cells. (D) Streptavidin-488 flow cytometry demonstrates that subtiligase-TM (red) more efficiently biotinylates cell surface N termini compared to soluble subtiligase added to cells (cyan). No enzyme control is shown in gray. (E) Streptavidin-488 flow cytometry shows that robust cell surface biotinylation is observed with activate subtiligase-TM (red) but not with the catalytically inactive C221A mutant (cyan). (F) A Western blot of subtiligase-TM-expressing HEK293T cells shows that biotinylation activity is dependent on both active subtiligase and the presence of a biotinylated peptide ester substrate. No biotinylation is observed when the inactive subtiligase-C221A-TM mutant is used or in the absence of biotinylated peptide ester substrate. (G) Fluorescence microscopy shows that subtiligase-TM expression (red) and biotinylation activity (green) are colocalized at the cell surface. No biotinylation activity is observed when the inactivate subtiligase-C221A-TM mutant is expressed.