Fig. 2 |. Glycosylation of Thr178 in FGF23 by GalNAc-T3 in cells requires its lectin domain activity.
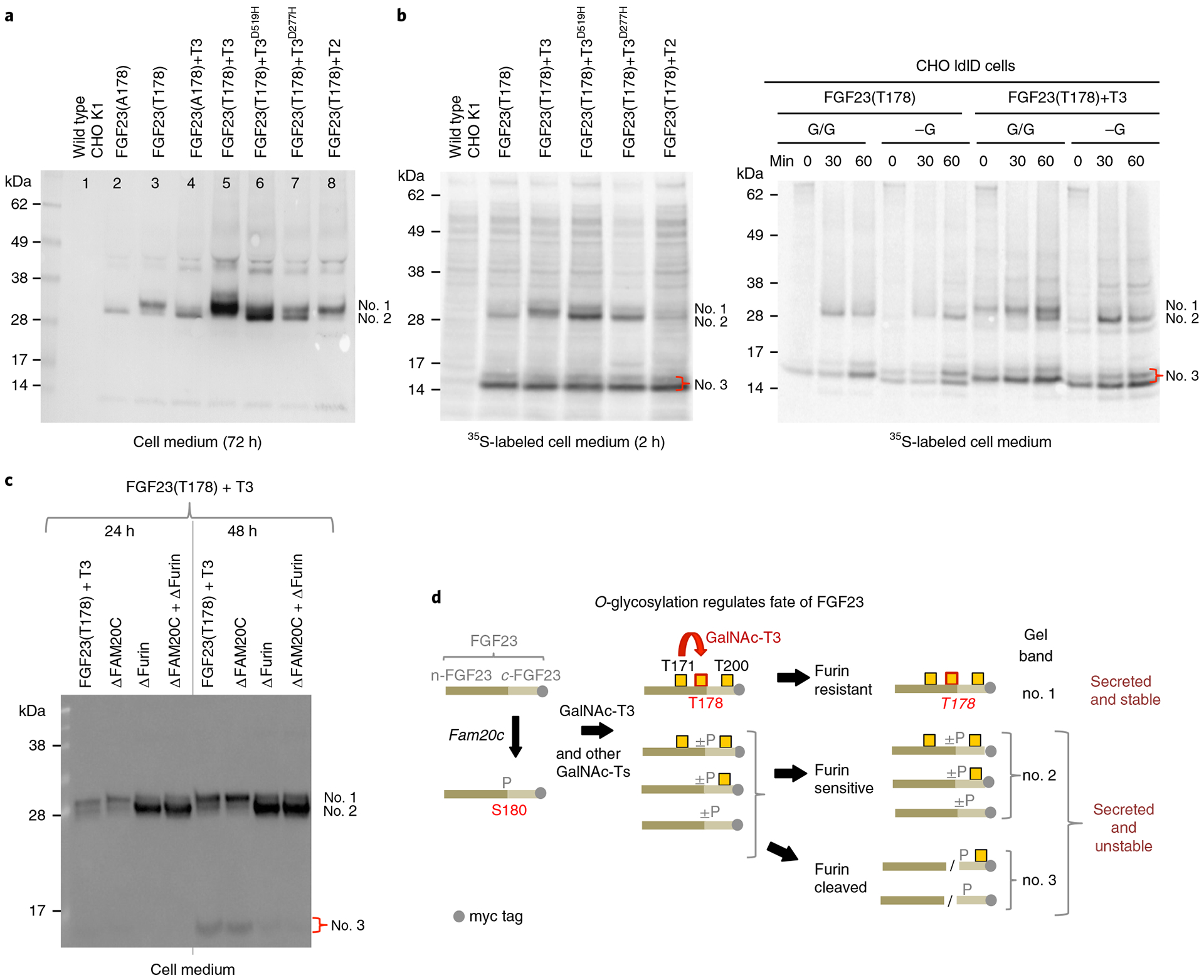
a, Anti-myc NuPAGE western blot analysis of culture medium of CHO K1 cells stably transfected with C-terminal myc labeled WT (FGF23T178) and mutant (FGF23A178) FGF23 with or without coexpression of WT and mutant GalNAc-T3 and WT GalNAc-T2. Medium was collected and analyzed after 72 h growth. The different glycosylation states of full length FGF23 are labeled: upper band (no. 1) FGF23 fully glycosylated at Thr178, lower band (no. 2) FGF23 lacking glycosylation at Thr178 but glycosylated at Thr171 and/or Thr200. b, Left panel shows NuPAGE analysis of 35S-Met labeled (2 h without chase) FGF23 (C-terminal myc tagged) expressed in CHO K1 cells with or without coexpression of WT and mutant GalNAc-T3 and with GalNAc-T2. Right panel shows NuPAGE analysis of 35S-Met labeled FGF23 expressed in CHO ldlD cells with or without coexpression of GalNAc-T3 under (G/G) or lacking (-G) conditions for O-glycosylation. The bands at (no. 3) represent the furin cleaved FGF23 C-terminal fragment. c, Western blot analysis of culture medium of CHO, CHO-ΔFAM20c, CHO-ΔFURIN and CHO-ΔFAM20c-ΔFURIN cells stably transfected with WT FGF23 and GalNAc-T3 demonstrating GalNAc-T3 only partially glycosylates FGF23 Thr178 under these conditions. d, Schematic representation of the possible fates of FGF23. Numbers to the right of the species correspond to the gel bands labeled in a–c. All experiments were performed in sextuplicate.
