Fig. 4 |. Crystal structures of TgGalNAc-T3 in ternary complexes with UDP-Mn+2-P3 and UDP-Mn+2-FGF23c.
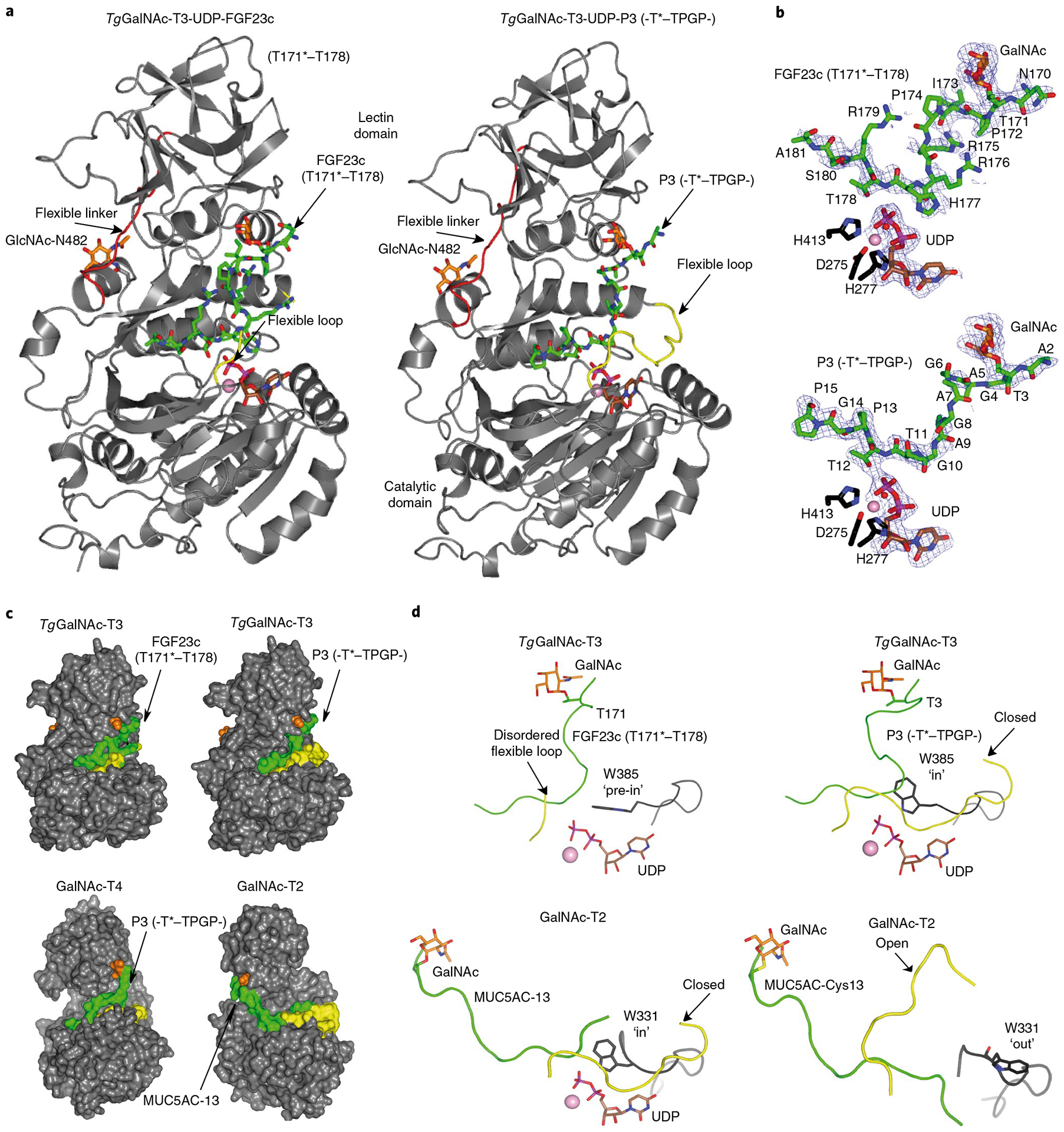
a, Ribbon structures of TgGalNAc-T3 ternary structures with glycopeptides FGF23c (left) and P3 (right). Note that the FGF23c glycopeptide is unstructured, resembling the unstructured region of the FGF23 C terminus within the alpha-Klotho–FGF23–FGFR1c complex25. b, Zoomed in structures showing the bound Mn+2, UDP and peptides (FGF23c, upper, and P3, lower). Electron density maps are Fo–Fc (blue) contoured at 2.2 σ for P3/FGF23c and UDP. Note that no electron density was visible for any of the Arg side chains and the EDDS sequence. In a and b the catalytic and lectin domains are colored in gray, the flexible linker and catalytic domain active site loops are depicted in red and yellow, respectively. The GalNAc moieties of P3 and FGF23c are shown as orange carbon atoms, while the remaining residues are shown as green carbon atoms. The UDP nucleotide is depicted with brown carbon atoms whereas the manganese atom is shown as a pink sphere. The remaining N-linked GlcNAc moiety bound to Asn482 is shown with orange carbon atoms. c, Surface representations of the complexes of GalNAc-T3–FGF23c–P3, GalNAc-T4–P3 (PDB 5NQA) and GalNAc-T2–MUC5AC-13 (PDB 5AJP) showing the different orientations of the lectin domain (compare GalNAc-T3/T4 with GalNAc-T2). The proteins, flexible loops, nucleotides, GalNAc moieties and peptide substrates are colored in gray, yellow, brown, orange and green, respectively. d, Expanded views of the TgGalNAc-T3–UDP–P3, TgGalNAc-T3–UDP–FGF23c, GalNAc-T2–UDP–MUC5AC-13 and GalNAc-T2–MUC5AC–Cys13 complexes showing their bound glycopeptide and the catalytic domain active site flexible loops. Note that Trp331 in GalNAc-T2–UDP–MUC5AC-13 and GalNAc-T2–MUC5AC–Cys13 complexes adopt ‘in’ and ‘out’ conformations, respectively, while Trp385 in the TgGalNAc-T3–UDP–P3 and TgGalNAc-T3–UDP–FGF23c complexes adopt in and ‘pre-in’ conformations, respectively.
