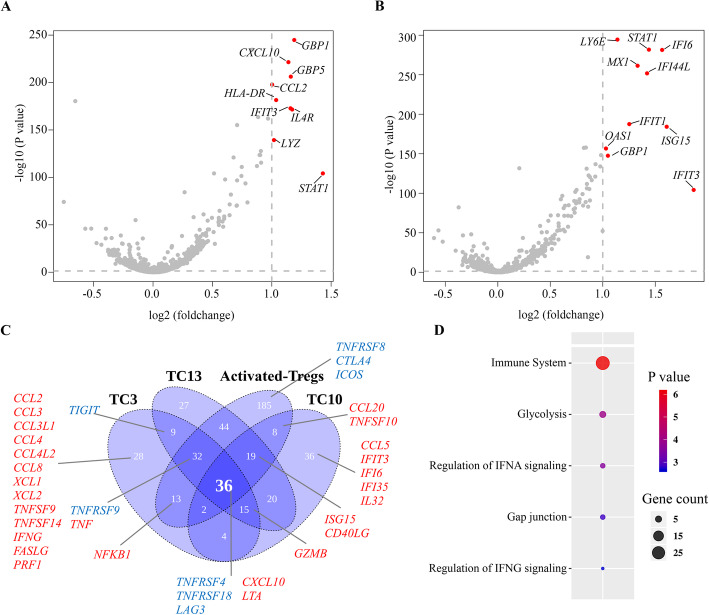
Fig. 3.
Blinatumomab-induced transcriptional changes and pathway analysis in T cell subclusters. a-b Volcano plot showing differentially expressed genes between a clusters TC5-CD4+ Naïve T and TC6-CD4+ Naïve T-STAT1, b clusters TC7-CD4+ TCM and TC8-CD4+ TCM-IFIT3. Genes with a P value < 0.05 and fold change > 2 are highlighted in red. c Venn plot showing the numbers of genes expressed differentially between the blinatumomab-activated and original clusters. Genes encoding cytokines and chemokines are labeled in red. Genes encoding co-signaling receptors are labeled in blue. TC3 represents the comparison between TC3-CD8+ Activated T and TC2-CD8+ TEM. TC13 represents the comparison between TC13-Activated T, TC0-CD4+ Naïve T, and TC5-CD4+ Naïve T. Activated-Tregs represents the comparison between Activated-Tregs and Resting-Tregs. TC10 represents the comparison between TC10-CD4+ Activated T and TC7-CD4+ TCM. d Pathway enrichment analysis revealed 36 differentially expressed genes listed in the reactome database (https://reactome.org/). The size of the circles is proportional to the Gene Count in the corresponding category. The color of each circle corresponds to the P value. Pathways are ranked according to their P value