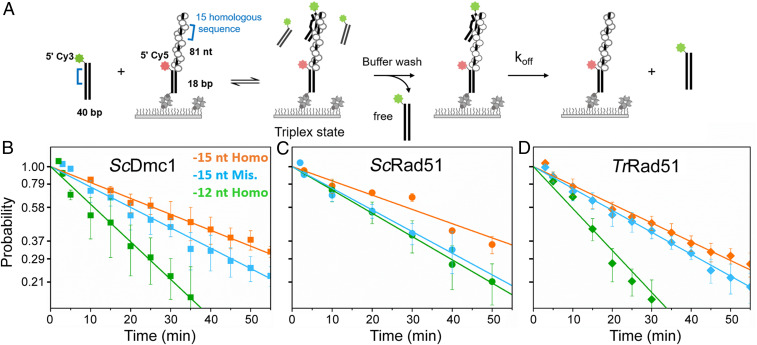
Fig. 5.
TrRad51 stabilizes mismatched triplex-state DNA. Single-molecule triplex-state stability experiments reveal similar stabilities of TrRad51 on sequences with full homology and sequences containing mismatched DNA. (A) Schematic of single-molecule triplex-state stability experiment. (B–D) Kinetics of triplex-state dissociation of 15-nt full homologous (in orange), 15-nt containing one mismatched sequence (in blue), and 12-nt full homology DNA substrates (in green) measured for ScDmc1 (B), ScRad51 (C), and TrRad51 (D), respectively. The proportions of triplex state DNA are plotted in natural log-scale, and the slope of the plot represents the dissociation rate. Error bars represent the SEM of three replicates. The dissociation rates are listed in SI Appendix, Table S17.