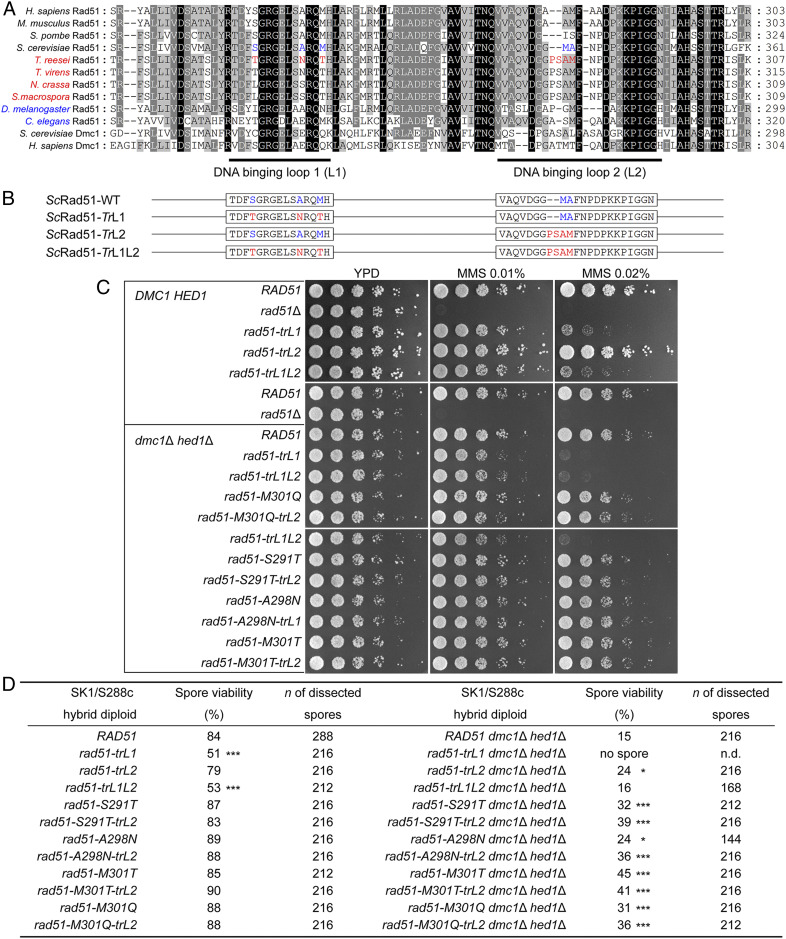
Fig. 6.
Chimeric ScRad51 with TrRad51 L2-specific amino acids can partially rescue the low spore viability phenotype of S288c/SK1 dmc1Δ hed1Δ hybrid diploid cells. (A) Comparison of Rad51 L1 and L2 amino acid sequences from “dual-RecA” and “Rad51-only” sexual eukaryotes. Species names in red indicate “Rad51-only” Pezizomycotina filamentous fungi; species names in blue indicate “Rad51-only” animals. L1/L2 amino acids that are ScRad51-specific and TrRad51-specific are highlighted in blue and red, respectively. Yeast and human Dmc1 were added to the alignment to aid comparison. Amino acids that are completely or partially conserved in these recombinase proteins are shaded in black or gray, respectively. (B) Schematic representation of S. cerevisiae mutant Rad51 proteins (ScRad51) carrying amino acid substitutions derived from T. reesei L1 (TrL1) and/or L2 (TrL2). Amino acids marked in red and blue represent residues with or without substitutions, respectively (SI Appendix, Table S18). (C) MMS resistance. Spot assay showing fivefold serial dilutions of indicated strains grown on YPD plates with 0, 0.01%, or 0.02% MMS. (D) Spore viability. Spore viability of indicated S288c/SK1 hybrid diploid cells was analyzed after 3 d on sporulation media at 30 °C. To score spore viability, only tetrads (but not dyads or triads) were dissected on YPD. Asterisks indicate spore viability of the rad51 mutants that is significantly different from that of WT under the same genetic background with P values calculated using Z-tests of two proportions (*P < .05 and ***P < .001). n.d. (not determined).