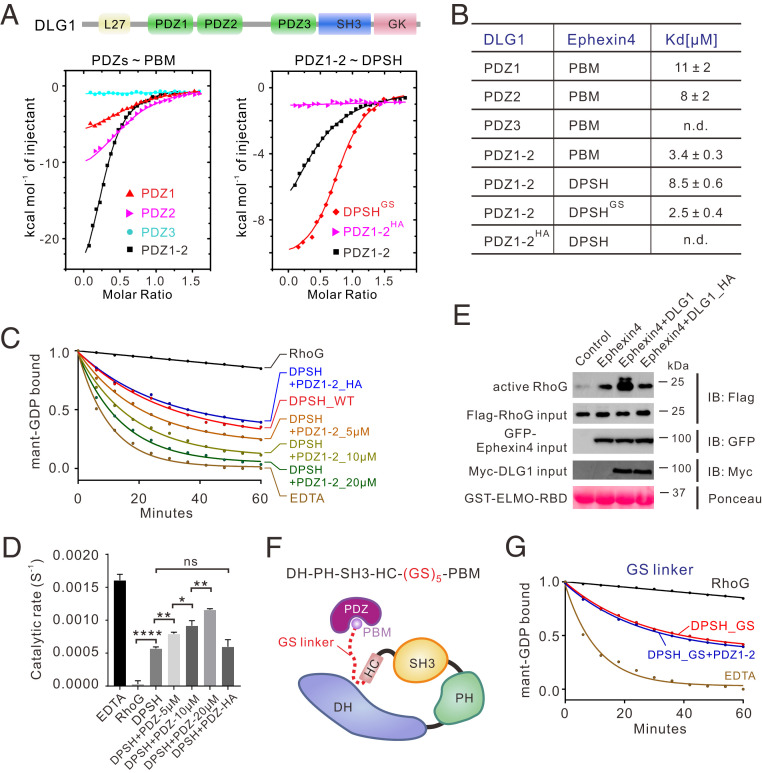
Fig. 3.
Binding of the PDZ domain relieves Ephexin4DPSH autoinhibition. (A) ITC curves of the bindings between WT or mutants of DLG PDZs and Ephexin4. (B) Summary of ITC-based measurements of binding affinities between WT or mutants of DLG PDZs and Ephexin4. n.d., not detectable. (C) Representative in vitro GEF assays showing that binding of DLG1 PDZ1-2, but not the PBM-binding deficient mutant, PDZ1-2HA, significantly enhanced GEF activity. (D) Quantification of GEF assays testing the role of the DLG1 PDZ domains in regulation of Ephexin4 activity shown in C. All results are expressed as mean ± SD. ns, not significant, ****P < 0.0001, **P < 0.01, *P < 0.1. (E) HEK 293T cells were transfected with the indicated plasmids. Active RhoG was coprecipitated with GST-ELMO2-RBD and detected by immunoblotting (IB). (F) Schematic diagram showing the engineering of a GS linker (GSGSGSGSGS) between HC and PBM in Ephexin4DPSH. (G) Representative in vitro GEF assays of DPSHGS with or without DLG1 PDZ1-2. Note that all GEF assays were performed using three independent protein preparations with at least duplicate measurements.