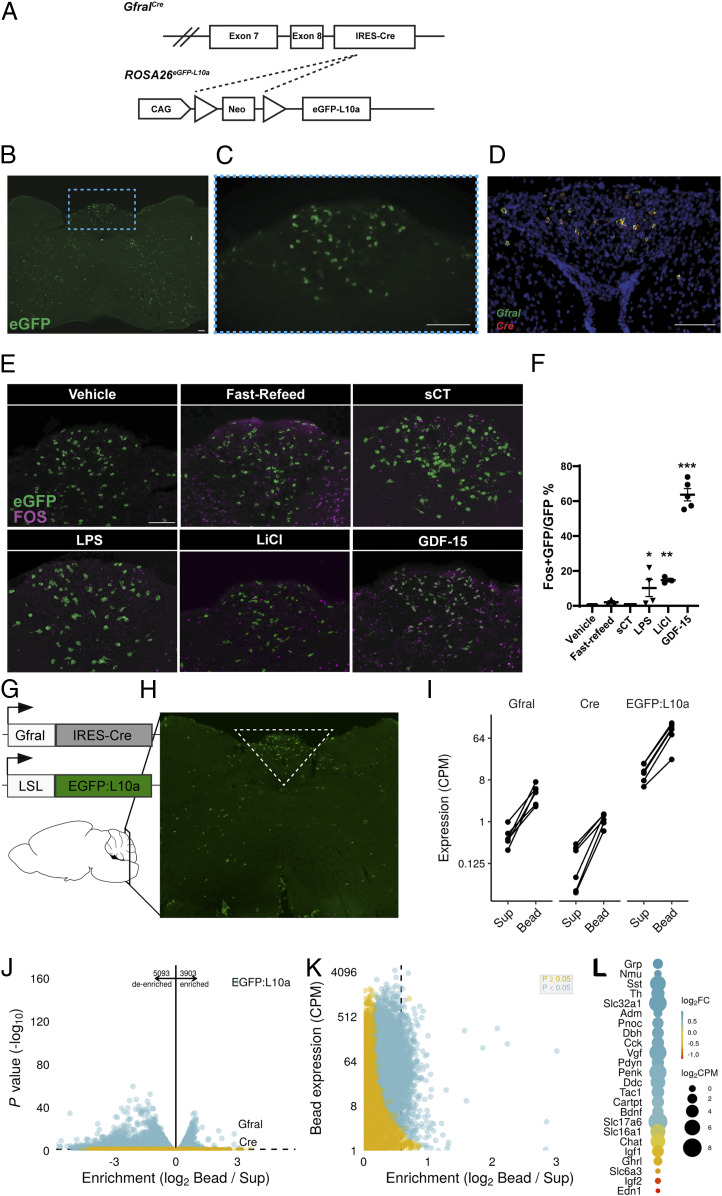
Fig. 1.
GDF-15 activates GFRAL neurons. (A) Schematic of the GfralCre-mediated excision of the Lox-Stop-Lox cassette from the ROSA26eGFP-L10a allele to generate GFRALeGFP mice. (B) Representative image of coronal hindbrain (Bregma -7.5) eGFP-IR (green) in GFRALeGFP mice. (C) High magnification of boxed AP region in B. (D) Representative image of ISH for Gfral (green) and Cre (red) transcripts in AP/NTS of GfralCre mice. (E and F) Representative image and quantification of FOS-IR (magenta) in GFRAL (eGFP+; green) neurons from GFRALeGFP mice in response to VEH, Refeeding, sCT, LPS, LiCl, and GDF-15 (n = 3–6). (G and H) Schematic of TRAP-Seq dissection of AP/NTS from GfralCre/+;Rosa26eGFP-L10a mice. (I) Expression of Gfral, Cre, and eGFP-L10a in Sup and immunoprecipitated actively translating ribosomes (bead). (J and K) Analysis of de-enriched and enriched transcripts within GFRAL neurons. (L) Assessment of neurotransmitter enrichment (Log2 Fold change gene expression in Bead compared with Sup) and expression levels in GFRAL neurons. Data are shown as mean ± SEM and analyzed with one-way ANOVA with Dunnet’s post hoc test (F), *P < 0.05, **P < 0.01, and ***P < 0.001. (Scale bar, 100 μm.)