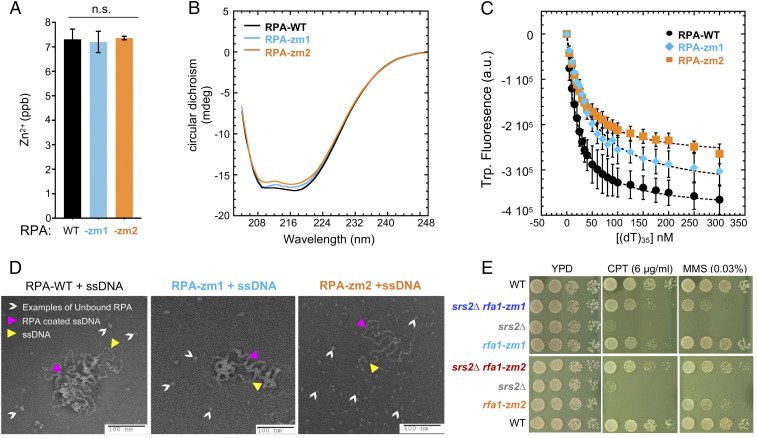
Fig. 2.
rfa1-zm mutants moderately reduce ssDNA binding and suppress srs2∆ genotoxin sensitivity. (A) ICP-MS results show that the concentration of Zn2+ per mole of RPA is similar between WT and mutant RPA complexes. (B) WT and mutant RPA complexes have similar CD spectra. (C) Intrinsic tryptophan fluorescence signals upon ssDNA binding by RPA complexes. (D) Negative stain electron microscopy images of RPA filaments on M13 ssDNA. RPA bound to ssDNA as nucleoprotein complexes are denoted by pink arrowheads. Examples of DNA-free RPA is denoted by white arrowheads. Examples of ssDNA region is marked with yellow arrowheads. More free RPA molecules were observed for RPA-zm1 and even more for RPA-zm2 compared to WT RPA. (E) rfa1-zm mutants suppress srs2Δ sensitivity toward CPT and MMS. Experiments were done as described in Fig. 1B.