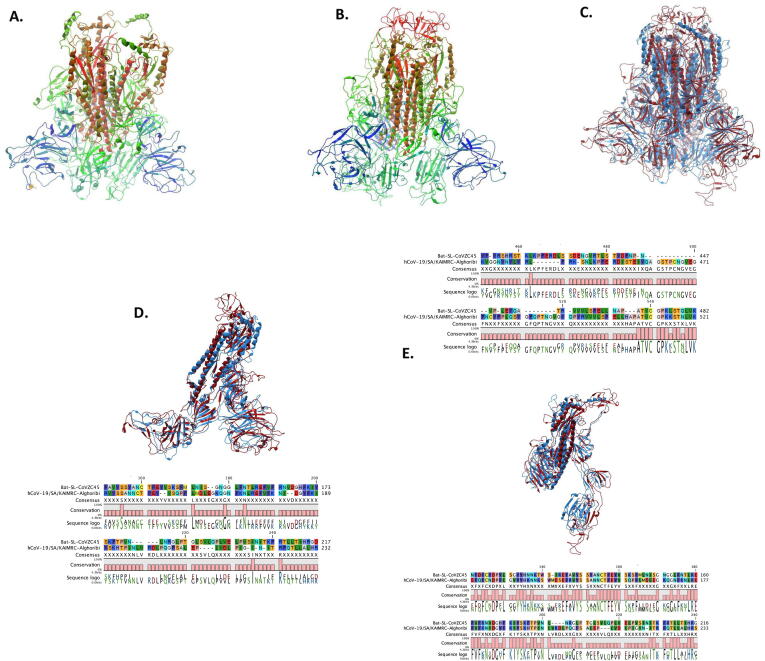
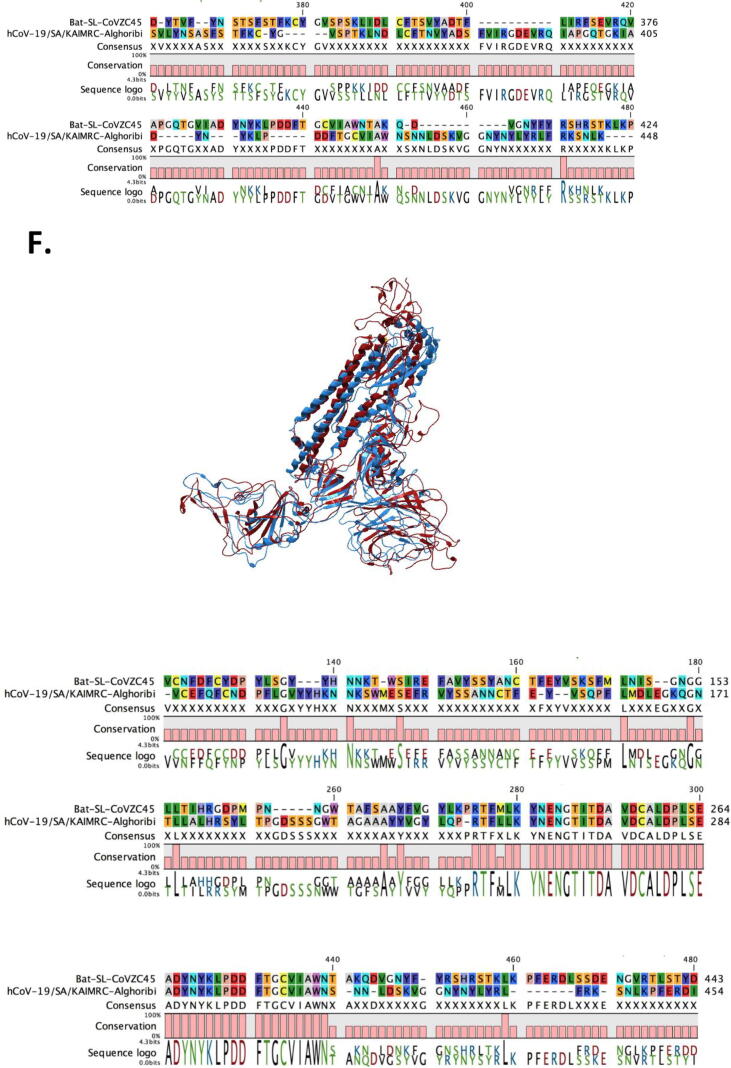
Fig. 2.
Spike glycoprotein homo-trimer structure modeling. SWISS model was used to get the best template and the fitting model (GQME = 0.75, QMEAN = -2.07, 0.94 coverage, 95% identity for hCoV-19/SA/KAIMRC-Alghoribi and GQME = 0.78, QMEAN = -3.87, 0.94 coverage and 76.73% identity for bat-ST-COVZC45) followed by importing the model to QIAGEN CLC Main Workbench V20.0 to obtain the final homo-trimer of (A) hCoV-19/SA/KAIMRC-Alghoribi and (B) bat-ST-COVZC45 and (C) models alignment. Three chains were observed for each of hCoV- 19 and bat-SL-CoV, thus each chain was aligned on basis of their amino acid sequence and their 3D structure, involving (D) Chain A, (E) Chain B and (F) Chain C.