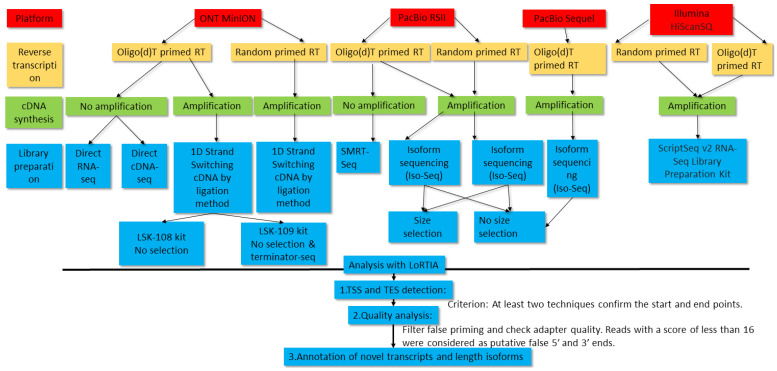
Figure 1.
Workflow of the PacBio, MinION, and Illumina sequencing. This data flow diagram shows the detailed overview of the study design. The LoRTIA program identified the transcription start site (TSS) and transcription end site (TES) positions in Oxford Nanopore Technologies (ONT) cDNA, direct cDNA (dcDNA), Terminator-seq, PacBio RSII random, IsoSeq, and Sequel samples. LoRTIA software suite also helped in the validation of TESs and introns in dRNA-Seq samples. The Illumina data were used for the validation of low-abundance transcripts, splice sites, antisense transcription, and for the identification of transcription readthroughs.