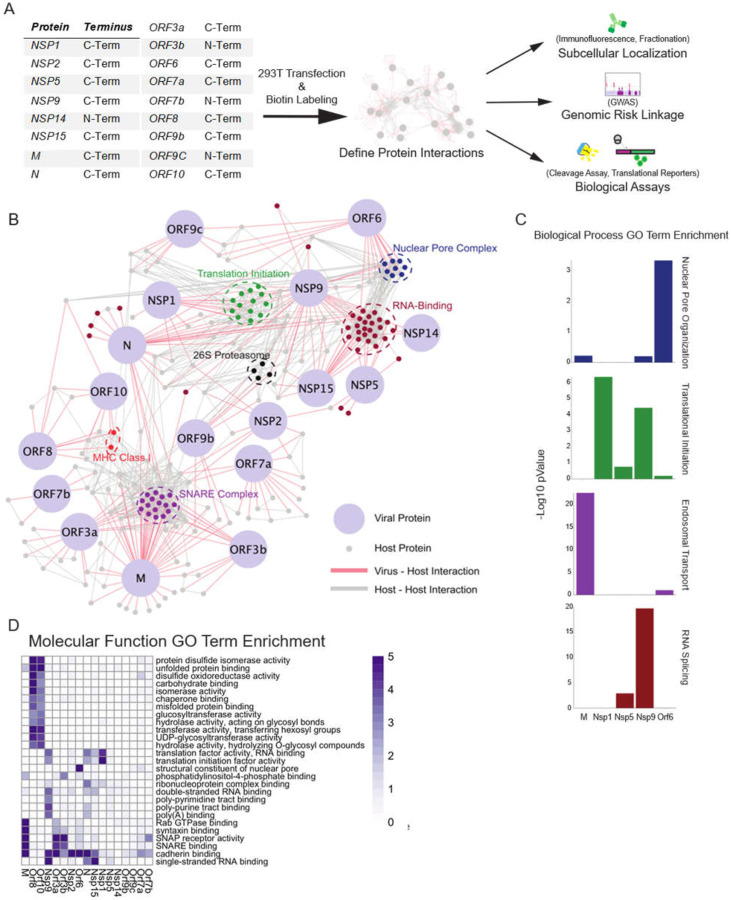
Fig. 1. Proximal Interactome of 17 SARS-CoV-2 proteins.
A) Schematic of BioID workflow. B) Curated network of SARS-CoV2 virus-host protein associations (SAINT ≥ 0.9) obtained from BASU BioID. Coronavirus proteins are labeled in light blue and virus-host interactions are connected by red edges, while host-host protein interactions obtained from high confidence STRING interactions are labeled in grey. Highlighted node clusters of similar function, including 26S proteasome components (black), MHC Class I (red), nuclear pore (dark blue), RNA-binding (maroon), SNARE complex (purple), translation initiation complex (green) proteins were selected based on GO term analysis. C) Selected biological process GO term enrichment; enrichment scores are given as -Log10 p-values. Selected GO terms are nuclear pore organization, translational initiation, endosomal transport, and RNA splicing. D) Heatmap of molecular function GO term enrichment of SARS-CoV-2 proteins. All presented GO terms have a -Log10 p-value >3 for the Nucleoprotein, the listed non-structural proteins, or the listed open reading frames or a -Log10 p-Value >5 for the M membrane protein.