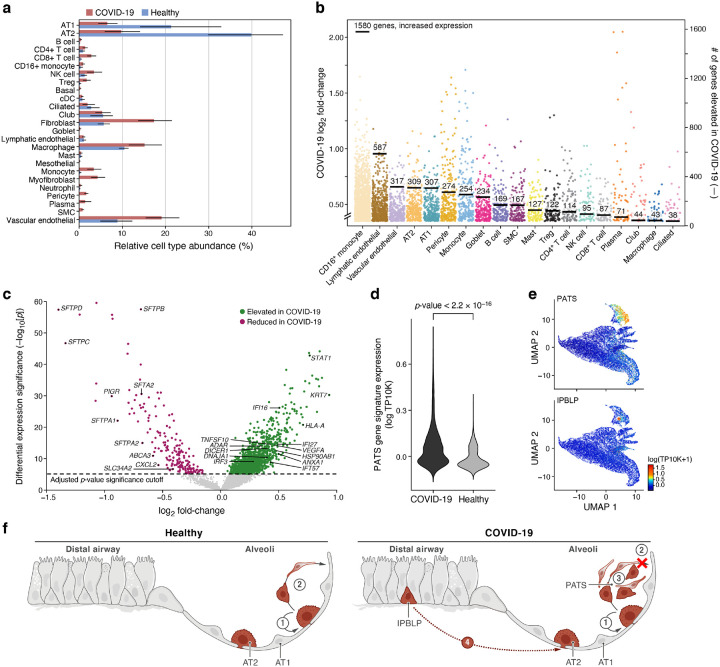
Figure 3. Dramatic remodeling of cell composition and cell intrinsic programs in COVID-19 lung.
a. Differences in cell composition between COVID-19 and healthy lung. Proportion (x axis, mean and 95% confidence intervals) of cells in each subset (y axis, by automatic annotation) in COVID-19 snRNA-Seq (red) and a healthy snRNA-Seq dataset (blue). b,c. Myeloid, endothelial and pneumocyte cells show substantial changes in cell intrinsic expression profiles in the COVID-19 lung. b. Log2(fold change) (y axis) between COVID-19 and healthy lung for each gene (dot) in each cell subset (x axis, by automatic annotation). Black bars: number of genes with significantly increased expression (adjusted p-value < 7.5*10−6). c. Significance (−Log10(P-value), y axis) magnitude (log2(fold-change), x axis) of differential expression of each gene (dots) in 2000 AT2 cells, from a meta-differential expression analysis between COVID-19 and healthy samples across 14 studies. d. An increased PATS37–39 program in pneumocytes in COVID-19 lung. Distribution of PATS signature scores (y axis) for the 17,655 cells from COVID-19 or 24,000 cells from healthy lung (x axis). e. UMAP embeddings of epithelial cells colored by their expression of cell program signatures (color legend, lower right) for the PATS program (upper panel) and the IPBLP program (lower panel). f. Graphical schematic of alveolar cellular turnover. In healthy alveoli (left panel), AT2 cells self-renew (1) and differentiate into AT1 (2). In COVID-19 alveoli (right panel), AT2 cell self-renewal (1) and AT1 differentiation (2) are inhibited, resulting in PATS accumulation (3) and recruitment of airway-derived IPBLP progenitors to alveoli (4).