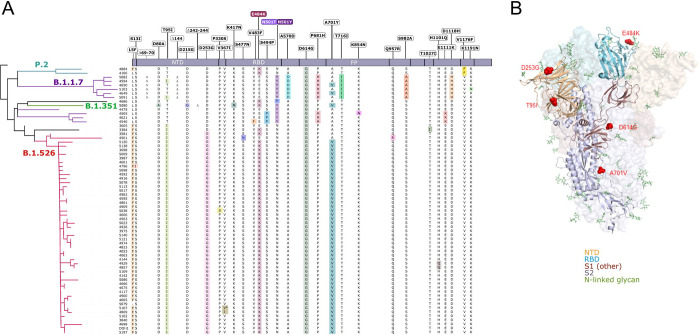
Figure 2. S-protein amino acid substitutions and structural changes represented in E484K- and N501Y-harboring isolates.
(A) Consensus sequences for 65 genomes were aligned against the Wuhan-Hu-1 reference genome (NC_045512). A phylogenetic reconstruction of these variant genomes is displayed on the left, generated using IQtree with 1,000 bootstrap replicates and maximum-likelihood tree search. Lineage assignments for major clades as determined by Pangolin are shown on the appropriate branches of the phylogenetic tree. On the right, residues at which at least one sample harbored a mutation compared to the reference genome are displayed above the S protein schematic. Amino acid substitutions identified in each sample are highlighted in color and wildtype residues are shown in black; an asterisk (*) indicates a silent nucleotide mutation. (B) Structural changes reflecting substitutions identified in the majority of B.1.526 isolates. Of the amino acid changes largely conserved in the B.1.526 lineage (L5F, T95I, D253G, E484K, D614G, A701V), D253G resides in the antigenic supersite within the N-terminal domain, a target for neutralizing antibodies, E484K at the RBD interface with the cellular receptor ACE2, and A701V near the furin cleavage site. Abbreviations – NTD – N-terminal domain; RBD – receptor binding domain; FP – fusion peptide.