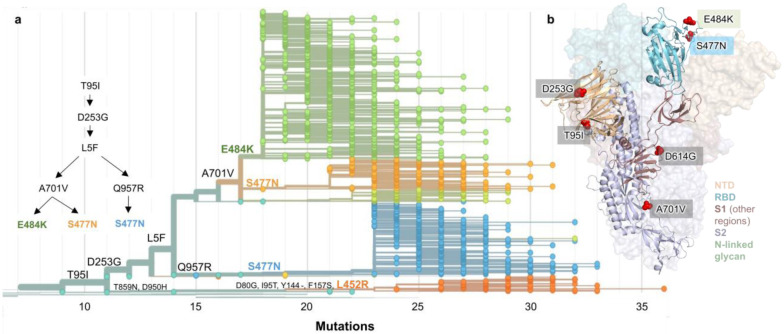
Figure 2. Spike protein amino acid substitutions and structural changes represented in sequenced isolates.
(a) Maximum-likelihood phylogenetic tree of 2,309 SARS-CoV-2 viruses colored according to spike protein haplotype. Spike protein mutations are labeled on the tree showing the stepwise accumulation of signature B.1.526 mutations T95I, D253G, and L5F, and branching of B.1.526-E484K (green) and two B.1.526-S477N sub-lineages (light orange, blue). The B.1.526-L452R sub-lineage (dark orange) emerged in parallel. An interactive version of this figure is available at https://nextstrain.org/groups/blab/ncov/ny/B.1.526. (b) Key mutations of B.1.526 displayed on the spike trimer. The D253G mutation resides in the antigenic supersite within the N-terminal domain (NTD), a target for neutralizing antibodies, E484K and S477N at the receptor binding domain (RBD) interface with the cellular receptor ACE2, and A701V near the furin cleavage site.