Figure 2: Epigenetic Modifications.
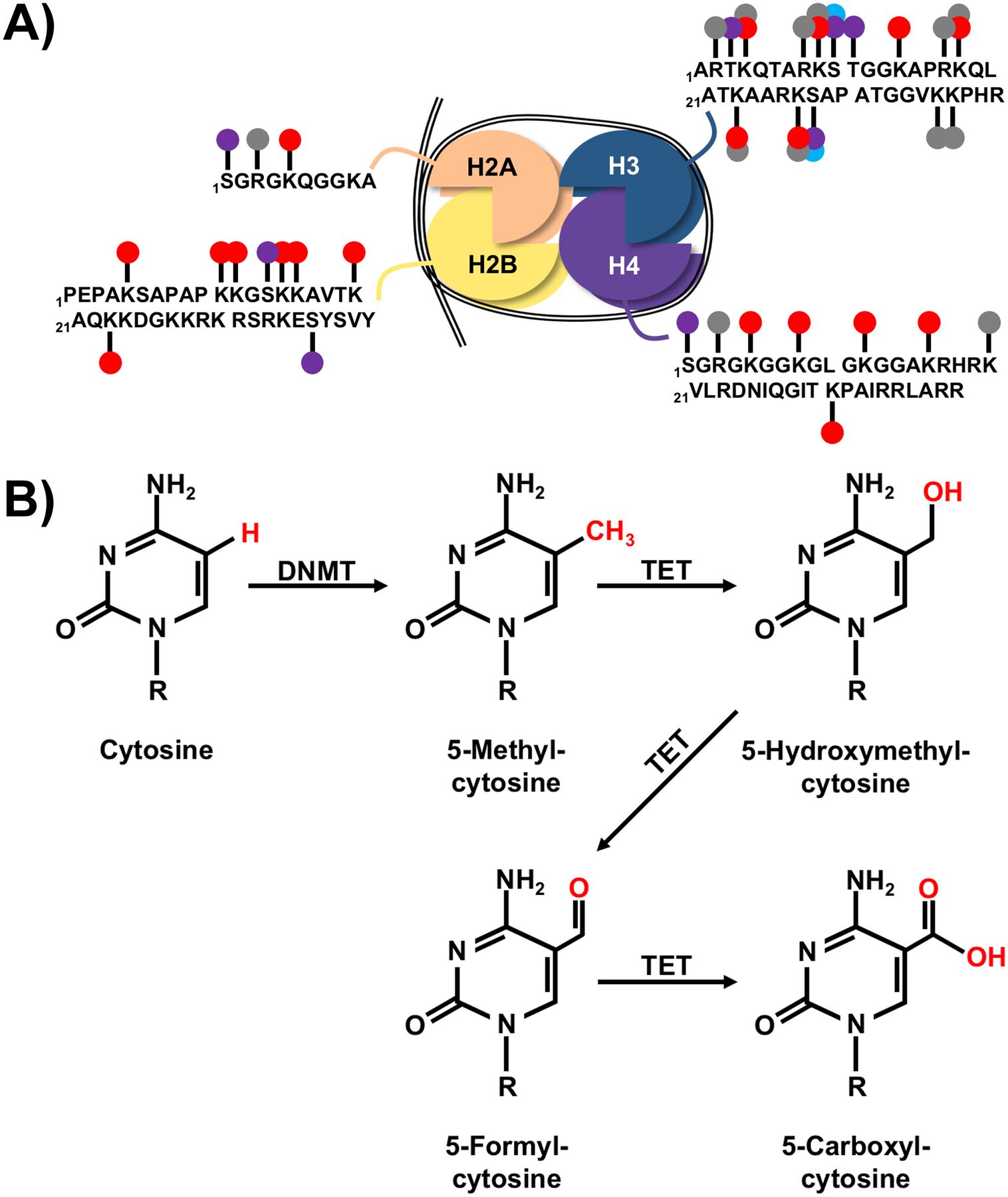
A) Nucleosomes are composed of an octamer of histones H2A, H2B, H3, and H4 (two dimers of 2A/2B and two dimers of H3/H4) with 146 base pairs of DNA wrapped around the complex (drawn as black double line). The H1 histone may also be attached as a linker to stabilize the DNA (not shown). Several epigenetically regulated modifications to the n-terminal tails of the histones influence DNA accessibility to transcription machinery including acetylation (red circles), methylation (gray circles), phosphorylation (purple circles), or ADP ribosylation (blue circles). The single letter abbreviation was used for each amino acid, with spaces after every ten, and subscript numbers indicating location. Multiple circles on a single amino acid represent alternative modifications. N-terminal sequences were based on consensus sequences for human histones at uniport.org: H2A - P0C0S8, H2B - P62807, H3 - P68431, H4 – P62805. B) DNA methyl transferases (DNMT) target cytosine residues immediately upstream of guanosine residues (CpG sites) within DNA to form 5-methylcytosine (5mC), which typically represses transcription. Ten-eleven-translocation (TET) proteins participate in demethylation by converting 5mC to 5-hydroxymethylcytosine (5hmC), which has also been demonstrated to participate in epigenetic regulation of gene expression. Further oxidation by TET proteins changes 5hmC to 5-formylcytosine (5fC) and subsequently 5-carboxylcytosine (5caC), which can then be converted back to cytosine (not pictured). The role of 5fC and 5caC in modulation of gene expression is uncertain due to the recent development of tools that can distinguish between 5mC, 5hmC, 5fC, and 5caC. For each structure, R is the sugar (deoxyribose)-phosphate group that forms the DNA nucleotide.
