Abstract
Radiomic analysis offers a powerful tool for the extraction of clinically relevant information from radiologic imaging. Radiomics can be used to predict patient outcome through automated high-throughput feature extraction, using large training cohorts to elucidate subtle relationships between image characteristics and disease status. However powerful, the data-driven nature of radiomics inherently offers no insight into the biological underpinnings of the observed relationships. Early radiomics work was dominated by analysis of semantic, radiologist-defined features and carried qualitative real-world meaning. Following the rapid developments and popularity of machine learning approaches, the field moved quickly toward high-throughput agnostic analyses, resulting in increasingly large feature sets. This trend took the focus toward an increase in predictive power and further away from a biological understanding of the findings. Such a disconnect between predictor model and biological meaning will inherently limit broad clinical translation. Efforts to reintroduce biological meaning into radiomics are gaining traction in the field with distinct emerging approaches available, including genomic correlates, local microscopic pathologic image textures, and macroscopic histopathologic marker expression. These methods are presented in this review, and their significance is discussed. The authors predict that following the increasing pressure for robust radiomics, biological validation will become a standard practice in the field, thus further cementing the role of the method in clinical decision making.
© RSNA, 2021
An earlier incorrect version appeared online. This article was corrected on February 10, 2021.
Summary
This review discusses the recent advances in the accelerated search for biological meaning of radiomics signatures, as the biological validation is gradually recognized as essential for the field to enter routine clinical practice.
Essentials
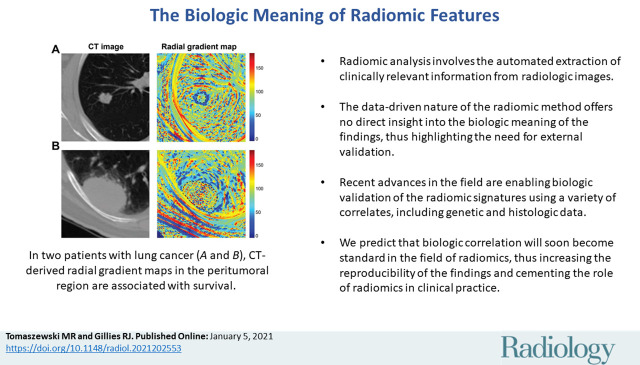
■ Radiomic analysis involves the automated extraction of clinically relevant information from radiologic images.
■ The data-driven nature of the radiomic method offers no direct insight into the biological meaning of the findings, thus highlighting the need for external validation.
■ Recent advances in the field are enabling biological validation of the radiomic signatures using a variety of correlates, including genetic and histologic data.
■ We predict that biological correlation will soon become standard in the field of radiomics, thus increasing the reproducibility of the findings and cementing the role of the method in clinical practice.
Introduction
Radiomics is an emerging field of research focused on the development of novel biomarkers based on data-driven analysis of radiologic images. It is predicated on the hypothesis that medical images reflect underlying pathophysiologic characteristics and hence, quantitative analyses can be useful to describe the biology of the imaged volume. Automated extraction of a large number of quantitative imaging features enables efficient elucidation of subtle characteristics within images that may be informative for disease diagnosis, prognosis, and treatment response. The development of advanced analytical and machine learning tools has led to rapid expansion of radiomics research and successful detection of patterns not available through qualitative radiologic analysis. Since it first appeared in print in 2012, publications referring to “Radiomics” have increased exponentially, numbering almost 1000 in 2019 (1).
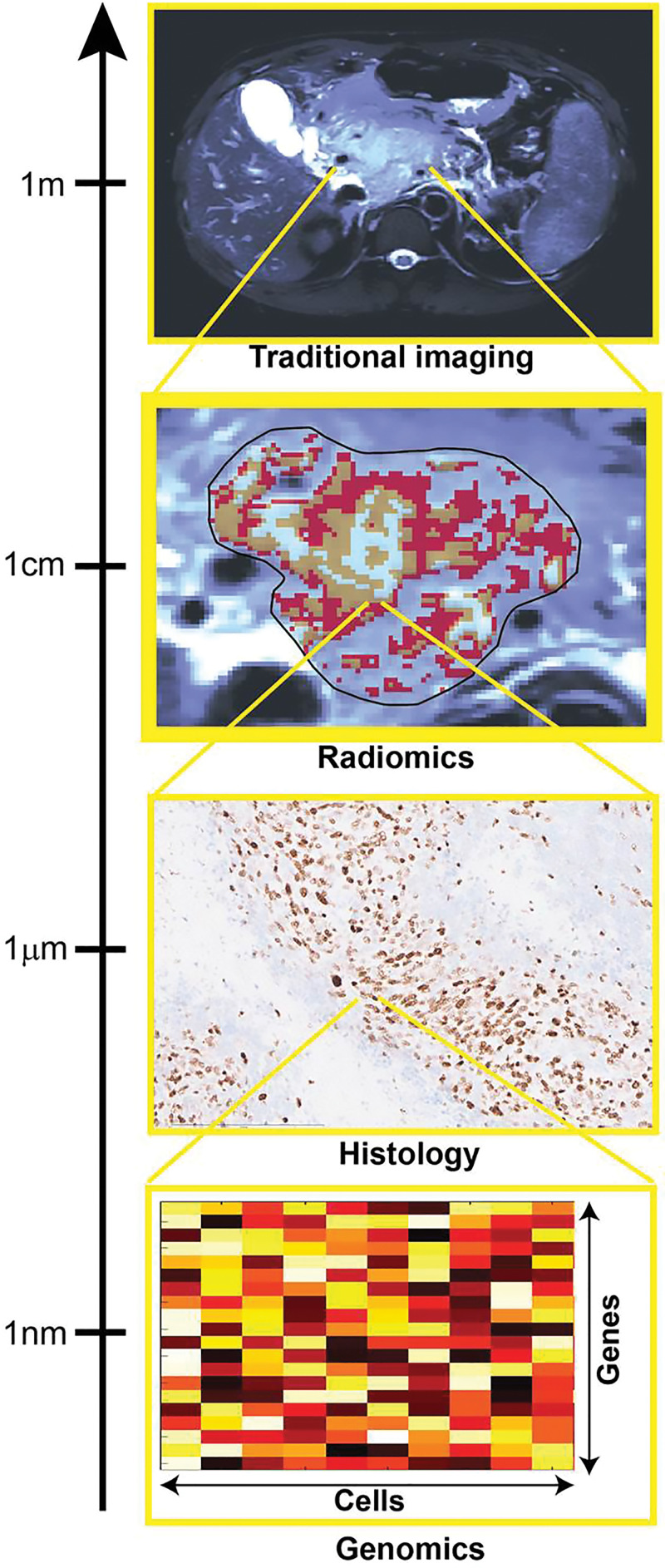
Modern medicine benefits from an enormous number of measurement techniques capable of informing disease characteristics that may not be accessible to physicians through manual examination. Biological processes can be tracked at spatial scales ranging from the level of the whole body all the way to single molecules, with imaging methods spanning most of that range. Radiomic analyses are predominantly based on anatomic and metabolic imaging, as shown in Figure 1. Through direct quantification of the tumor imaging phenotype at the spatial scale within the resolution of the imaging technique used, radiomics aims to provide surrogate, indirect insight into multiple aspects of the disease, including tumor grade, histologic and genetic subtype, and predicted outcome. These characteristics are reflective of alterations occurring at different spatial scales to the data provided by radiomics. Hence, the biological basis of the indirect relationships enabling radiomic predictions remains largely unexplained in most studies. Importantly, as the radiomic data can be captured longitudinally, it can be used to quantify the response of the underlying “biome” to external perturbations.
Figure 1:
Image shows how multiscale quantification provides complementary tumor insight. Histologic and genomic analysis can provide specific small-scale insight useful for validation of radiomic results, focused on quantification of spatial patterns of size exceeding image resolution.
Radiomic biomarker development is almost entirely data driven. In contrast, traditional biomarker development is generally driven ab initio by biology-based hypotheses. Preclinical experiments can often enable validation of the mechanism of action and informational content of the developed metric before translation and verification of clinical use. Approaches such as genomics, transcriptomics, proteomics, and radiomics are used to screen large-parameter spaces to find sensitive markers for prediction of outcome and thus often involve posthoc generation of hypotheses. Without an underlying biological rationale, the black box–like nature of “omics” methods significantly hinders its wider use and makes validation particularly challenging. Providing the biological context of the informative radiomics features will constitute an important step toward general acceptance of radiomics as a standalone diagnostic, predictive, or prognostic method in the radiology and oncology communities. As the field develops and joins the mainstream, the emerging radiomic signatures will need to adopt the reporting guidelines (2) and evaluation standards, as used by other novel clinical diagnostic and prognostic approaches, including comparison to existing reference standard methods. Biological validation will be essential as part of this process.
In addition, understanding of the biological underpinning of the observed relationships, where possible, can strengthen the conclusions and can provide additional validation and opportunities for investigation. For example, if a highly prognostic radiomic feature were found to be highly correlated to the expression of a particular protein, one could then investigate the relationship between the protein and the outcome. This review discusses the studies constituting notable developments in biological validation of radiomic findings, as perceived by the authors, and postulates the implementation of new standards for validation, thus prioritizing the efforts to establish a biological basis of the findings.
Importance of Validation
Radiomics is intrinsically data-driven by screening a high volume of features for reproducibility and information content. However, because many features are considered, there is a real danger of overfitting or overinterpreting the derived models. Together with increased sophistication of the analytics, more stringent validation of the findings is increasingly required. Most published radiomics studies present no validation of the proposed signatures beyond the use of an independent test set. This approach does not address much of the critique concerning the practical value of the findings because the causal relationship between radiomics and outcome may remain unclear. This limitation has been recognized in the development of a “radiomics quality score,” which is based on different evaluative metrics during construction of radiomic models (3). Key to this are the standardization of feature extraction and statistical approaches (4), the use of separate validation cohorts from multiple institutions (5), and an increasing involvement of radiomics in prospective clinical studies (6). Further efforts for standardization and streamlining of the radiomic analysis process are led by the Radiomic Ontology project (7), which provides a comprehensive analytical platform for clinical use, including solutions for multicenter studies (8). Although these approaches contribute to improved reproducibility and impact of the findings, biological validation remains a critically important and elusive metric and is necessary to transform radiomic analysis into an actionable part of the routine clinical decision process.
The level of biological insight and tools used in other high-throughput data-driven studies are largely not available for radiomics. For example, the genes and pathways identified in genomics screenings can be interrogated in vitro and, thanks to the large body of research available, embodied into powerful resources such as the Gene Ontology (9), which combines the functional information available on each gene and pathway. We believe that as the field of radiomics grows, and more information related to biological underpinning of different features becomes available, a similar database may be compiled and published. Stringent standardization of the analytical methods, as described herein, will be paramount for this task.
In this review, the major efforts for biological validation of radiomic findings are presented and are divided into main sections focused on the main classes of biological correlates available for comparison.
The relationship between data-driven radiomics and visual image content, described by semantic features, is first discussed, followed by separate sections describing the biological insight available for radiomics validation from the notable correlates—genetic and histopathologic data. Finally, habitat imaging is discussed as a radiomic approach that holds significant promise for effective validation of the biological findings underlying the image heterogeneity.
The review aims to summarize and discuss the approaches available and practiced in the field for biological validation to promote their more widespread use. Although many studies present relationships between radiomic features and biological correlates, such as histologic tumor grade or gene expression, emphasis in this review is placed on reports where additional independent correlates were discussed to validate the biological source of the findings. We strongly believe that introduction of biological validation into standard practice for radiomic model building will accelerate clinical acceptance and routine use of the method in patient care.
Semantic Analyses
Semantic features are generally radiologist-defined accepted metrics that describe tumor morphologic characteristics and location. Examples include spiculation, lepidic, concavities, and central necrosis, among other terms. Before the use of more sophisticated approaches and data-driven features, early work in the field of radiomics can be associated with combining multiple semantic features (10) into more complex signatures (eg, for gene expression prediction) (11,12). The visual nature of the considered characteristics (13) ensured the findings remained relatively grounded in physical, if not biological, understanding. Following the rapid development and popularity of computer vision and machine learning approaches, the field of radiomics moved quickly toward high-throughput agnostic analysis and complex combined signatures. This led to increased throughput and lower cost of the analysis, at the price of biological understanding.
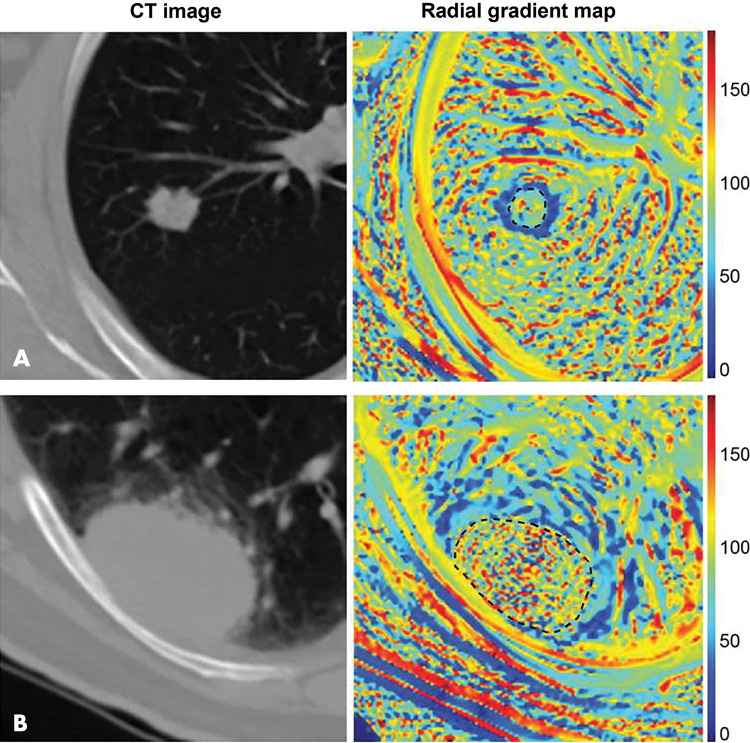
Some notable recent studies including semantic and conventional (agnostic) image features suggest that these approaches may provide valuable intuitive and biological insight, as they clearly relate to the visual phenotype. Yip et al (14) quantified the correlations between manual semantic features and automatically computed radiomic features in non–small cell lung cancer, showing relationships between the two approaches. Importantly, the authors used the observed relationships to discuss the physical and biological source of radiomic feature changes. Similarly, Tunali et al (15) introduced an automated framework for robust quantification of the tumor-stromal interface features, showing its association with survival in non–small cell lung cancer. The visual interpretation of the novel features was then verified by showing significant correlation to semantically scored features such as lobulation and border definition. An example of this quantitative approach is shown in Figure 2. The introduction of decision trees into model design may also simplify intuitive understanding of the features’ informational content. Gevaert et al (16) used this approach for the successful prediction of mutation status using semantic and computed features. The authors highlight, however, an important shortcoming of the semantic analysis—its poor interobserver reproducibility related to subjective assessment, expressed in low Cohen κ.
Figure 2:
Radiomics can quantify visual tumor characteristics. CT images (left) of lung cancer lesions from two patients (A and B) were used by Tunali et al to calculate radial gradient maps (right) that describe tumor edge interface (tumor outlined in black dotted line). Quantification of simple mean and standard deviation of map in peritumoral region is associated with survival and correlates to qualitative semantic descriptors of tumor edge, such as border definition. Source.—Reference 15.
Some studies have focused on automated quantification of particular visual characteristics of the tumor to improve reproducibility and standardization of the analysis. Koay et al (17,18) focused on quantitative assessment of the visual conspicuity of pancreatic tumors in CT images, presenting its power for survival prediction. This approach could then be evaluated against histologic and genomic data to shed more light on the microscopic characteristics underlying the measured phenotype. This hypothesis-driven study employs a few custom image features in place of a standard radiomic approach, which, when feasible, may be preferable for identification of an unambiguous relationship between the image-based and biological information.
Similarly, multiple quantitative metrics of tumor heterogeneity have been proposed, as detailed in a review by Davnall et al (19). Notably, only a small proportion of studies attempting to quantify heterogeneity discuss the informational content of the chosen metrics, with important examples coming from dynamic contrast-enhanced MRI (20,21) and fluorine 18 fluorodeoxyglucose PET (22). Most other studies instead consider panels of textures, such as gray-level co-occurrence matrix radiomic features, which describe the internal image intensity patterns, assuming an implicit relationship between these and heterogeneity. Although relatively little data are available on the relationships between the qualitative definition of heterogeneity and image texture features, some studies report indirect relationships. Skogen et al (23) demonstrated in glioma that tumor grade, known to be associated with visual heterogeneity (24), correlated to a standard deviation of intensity distributions in filtered CT images. Conversely, Liu et al (25) reported no correlation between tumor grade and visual heterogeneity, as assessed manually, whereas texture features commonly associated with tumor heterogeneity showed a strong link. More studies validating the relationships between the visual characteristics, semantic features, and quantitative metrics assumed to represent them may be required.
Radiogenomics Relationship with Gene Expression
Data-driven image feature extraction can also be combined with genetic analysis to inform mutation status beyond survival and tumor grade prediction. This is referred to as radiogenomics, not to be confused with the same term used to define relationships between genomics and radiosensitivity. Radiogenomics is a rapidly developing high-throughput method aimed at extracting and correlating multiple image features with genomic information. It is increasingly accessible thanks to The Cancer Genome Atlas initiative (26), in particular in combination with the resources offered by associated The Cancer Imaging Archive (27). Although mainly focused on providing surrogate imaging signatures for genetic information, radiogenomic (28) tools can also provide some biological validation of radiomic signatures. The similarities between the analytical approaches used in feature extraction from radiologic and genetic data can also be exploited (29) for cross-validation between the methods. In a comprehensive study of the relationship between somatic mutations and radiographic CT phenotype in lung cancer, Rios Velazquez et al (30) showed significant associations between multiple radiomic features and several relevant mutations, including epidermal growth factor receptor, or EGFR, and Kirsten rat sarcoma viral oncogene homolog, or KRAS, in non–small cell lung cancers. Aerts et al (31) used genetic information to shed light on the biological characteristics of a developed radiomic signature of survival in lung and head and neck cancers. Conversely, Gevaert et al (32) used the genetic signatures as a starting point to predict image features, thus further reinforcing the strong link between genomic and radiomic information.
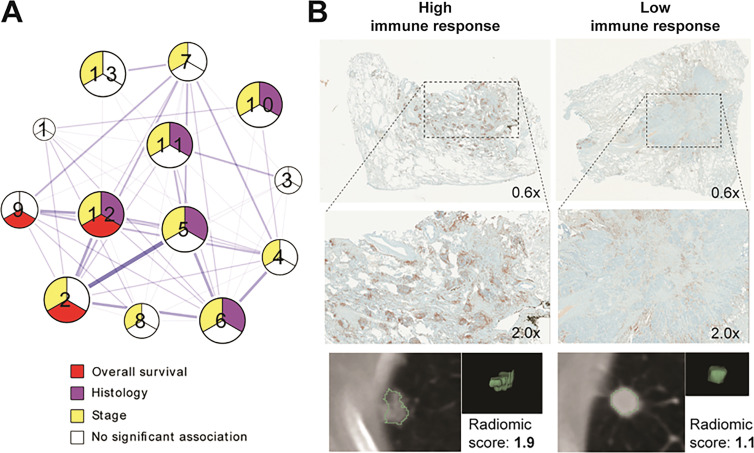
Epidermal growth factor receptor status in non–small cell lung cancer is a widely researched subject because of its high frequency and the fact that this mutation is actionable for treatment with tyrosine kinase inhibitors (33,34). Multiple studies reported CT signatures associated with epidermal growth factor receptor mutation status as determined with genetic testing. As pointed out by Yip et al (35), many of them have, however, shown conflicting conclusions (36–38) that undermine the reproducibility of the approach. Combining the genomic correlations with other biological metrics as provided by histologic analysis may prove necessary to ultimately verify such inconsistent findings. In a study following these principles, Sun et al (39) developed a radiomic signature of immune infiltration, relating the score to relevant gene expression panel, pathologic findings, and survival data. Similarly, Tunali et al (40) demonstrated that the CT radiomic features that were most predictive of survival following immunotherapy were also strongly associated with hypoxia according to genetic profiling and immunohistochemistry (IHC). Mu et al (41,42) have used PET/CT radiomics to develop predictors of both programmed death-ligand 1, or PD-L1, status and epidermal growth factor receptor mutation status. When combined, these generated a powerful decision support tool, as these two phenotypes are generally mutually exclusive. Beyond additional validation, the combination of multiple correlates in these studies informs on the biological driving forces of the radiomic relationships. Beig et al (43) took advantage of the known relevance of oxygenation status in cancer by developing a radiomic signature of tumor hypoxia from patient MRI scans and subsequently showing it to relate strongly to the survival of patients with glioblastoma. Grossman et al (44) presented an alternative approach with the aim to reveal general themes in the relationships between radiomic feature classes and pathway information. A biclustering approach was used to identify clusters of radiogenomic correlations, linked to outcome, which were subsequently validated to show, for example, correlations with immune infiltration or nuclear factor-κB, or NF-κB, expression (also involved in immune response), with targeted IHC (Fig 3). Introduction of histologic information can provide the necessary link to understand the relationships between genetics and seemingly distant radiomic features describing the macroscopic tumor textures.
Figure 3:
Associations between radiomic and pathway data can be explored histologically. A, Diagram depicts analysis of correlations between pathway enrichment and radiomic features presented by Grossman et al. Multiple clusters (numbers 1–13) describe relationships between distinct biological processes and image information. Further correlation of relevant radiomic features and immunohistochemical staining (nuclear CD3 expression) was performed to validate findings and to provide link to understand interaction between genetics and imaging characteristics of tumor. B, Example tumors with strong immune response and/or high radiomic score (left) and low immune response and/or radiomic score (right) are shown. Source.—Reference 44.
Instead of considering the tumor volume, Wu et al (45) focused on the image features of tumor-adjacent parenchyma and concluded that their predictive power for patient survival was likely associated with the related dysregulation of relevant signaling pathways, such as tumor necrosis factor-α, or TNF-α. Modules of features of similar informational content were grouped and related first to genetic data for screening to later evaluate their power for survival prediction. In a similar study, this same group performed an analysis of breast cancer, demonstrating the use of radiomic analysis of contrast-enhanced MRI to identify tumor subtypes of distinct survival and molecular pathway characteristics, likely underlying the survival differences (46). Itakura et al (47) took a distinct approach by using unsupervised analysis to define main clusters of tumors, defined strongly by their shape and enhancement pattern. Following confirmation of differential survival of the clusters, corresponding genetic pathway upregulation differences were identified, providing potential biological underpinning of the outcome disparity.
Treatment response information can further verify the biological mechanism behind a radiomic model. Liu et al (48) used MRI perfusion imaging data to identify subgroups of patients with glioblastoma with distinct survival. This was further interrogated with associated genomic data to relate contrast enhancement to angiogenic properties of the tumor, and the model was shown to correctly predict response to antiangiogenic treatment, confirming its biological underpinning.
The relationships observed in radiogenomic studies may often still be biologically ambiguous, given the complexity of the genetic code and its link to the phenotype, which is often indirect. Additionally, the large number of correlated parameters, both image- and genome-derived, results in an often overwhelming number of comparisons, which can blur conclusions.
Demonstration of a causal relationship between tumor characteristics and image features remains beyond the reach of most radiomic studies, as the predominantly clinical and retrospective nature of the work precludes an intervention to test observed correlations. This may be informed by preclinical studies. Panth et al (49) argued causality between genetic changes and radiomic features with the aid of an inducible gene mouse model, shown to affect specifically tumor hypoxia. In a broader coclinical study, Zinn et al (50) demonstrated a causal link between gene expression and radiomic signature changes through analysis of wild type versus knockdown mouse tumor models and related these to patient data. These studies offer more detailed and powerful insight into the mechanism of action and biological underpinning of the findings and should therefore be encouraged.
Additional Validation of Histopathologic Findings and IHC
One step closer to the anatomy is the histopathologic information describing the phenotypes of the tumor cell population and its surrounding microenvironment. Establishing the relationship between observed radiomic signatures and pathologic findings could provide a powerful link to the biological drivers of patient outcomes, potentially more specific than genetic profiles.
Multiple studies have used histologic data to provide more detailed end point information compared with standard survival metrics, thus demonstrating the potential for radiomics to predict, for example, the histologic tumor type (51–53) or pathologic response (54,55). Others focused on prediction of yet more specific analysis, for example, with Yin et al (56) relating PET/MRI radiomics to vascular density derived from pathologic samples. This study did not, however, use the biological correlates available from histologic data for validation and improvement of radiomic findings. In a novel approach to pathologic finding–supported radiomic model development, Tang et al (57) stratified patients according to two immune-pathologic markers and used this as an additional screening step. They arrived at a survival predictor correlated to the relevant immunologic phenotype, known to be relevant for patient outcome.
Sun et al (39) have used IHC staining information to validate the biological underpinnings of a proposed survival signature, which was related to immune infiltration. Ha et al (58) demonstrated the use of histopathologic correlations in evaluation of unsupervised radiomic models. The clusters of tumors based on radiomic features were shown to display differential response to treatment and recurrence risk. They were also observed to present differential expression of relevant IHC markers, thus providing an insight into the probable biological source of the difference in outcomes.
Biological validation of the mechanism linking a radiomic signature to patient outcome requires careful selection of a meaningful correlate. Sone radiomic studies have focused on relating the signatures to tumor oxygenation status (hypoxia), measured through association to relevant genetic pathways, as described earlier, and/or with the aid of histopathologic analysis, relying on the known strong link between tumor hypoxia, disease progression, and treatment response across multiple cancer types (59,60). Tunali et al (40) confirmed a relationship between the radiomic model of survival in non–small cell lung cancer and IHC of a hypoxia marker, carbonic anhydrase IX, which was originally identified in a radiogenomic screening. Similar to Sun et al (39), IHC analysis was used to bridge the imaging and genetic information, thus overcoming the limited specificity of pathway data to direct phenotypical insight.
Conversely, radiomic approaches can be used to provide surrogate measure of tumor hypoxia. Traditionally, direct tumor hypoxia measurements (61) are attempted with molecular imaging techniques, including specific PET and optical probes (62,63). Although novel tracers are being developed (64), the imaging efforts have largely been hampered with low dynamic range and hence poor signal-to-noise ratio. In contrast, radiomic analyses of standard of care imaging have a potential to provide an indirect insight into the hypoxia status. As described previously, Beig et al (43) used a radiogenomic signature of hypoxia to predict survival in patients with glioblastoma. Pimonidazole sequesters in hypoxic volumes and can be detected with IHC. A data-driven analysis relating pimonidazole staining to tumor characteristics has previously been described using genomics (65). Ganeshan et al (66) took a more detailed look at the image texture features and histologic metrics of hypoxia and angiogenesis in coregistered radiologic and IHC images and reported multiple significant associations.
Muzi et al (67) and Sörensen et al (68) developed moderately strong survival signatures based on image feature extraction from fluorine 18 fluoromisonidazole PET uptake imaging related directly to tumor hypoxia information. No secondary correlates were reported. As fluorine 18 fluoromisonidazole PET is not widely available to patients, Crispin-Ortzuar et al (69) have developed a surrogate signature of this PET hypoxia marker using the traditional FDG/PET and CT data. Another study by de Jong et al (70) describes the use of PET/CT radiomics in an attempt to measure changes in response to a hypoxia-altering nitroglycerin treatment. No significant differences were reported.
Local Radiomic Analysis Using Pathologic Correlates
A separate approach to relate radiomic results to tumor pathologic findings relies on texture analysis of histologic images. The emerging and rapidly expanding field of pathomics (71) aims to apply high-throughput image feature extraction techniques to interrogate the microscopic patterns in pathologic data, especially from hematoxylin-eosin–stained sections. Because of the close similarity of the approaches, the features from in vivo images may be compared with the features extracted from ex vivo specimens, often benefiting from a clearer biological definition of the image patterns and hence a better understanding of the features. The quantitative analysis of histologic data has been shown to improve outcome prediction (72,73) and to aid prognosis (74) beyond the capabilities of human practitioners, mimicking the goals of radiomics. Saltz et al (75) argued that the similarities between radiomic and pathomic analysis renders the combination of the techniques promising to improve the predictive power. Direct application of radiomic tools to histopathologic images has shown promise for tumor staging (76).
Comparing the radiomic features derived from macroscopic resolution in vivo images to subcellular scale data of pathomics is a challenge and may not provide direct insight into the biological underpinning of radiomics. As with the radiogenomics analysis, a significant gap in the biological source of information from the two approaches precludes clear conclusions. In a promising early attempt, Geady et al (77) generated simulated CT images from pathologic data, thus showing good correlations between the underlying microscopic information and derived texture features.
Other approaches have been proposed to overcome this limitation by focusing on histologic information at spatial scales that were matched to the in vivo imaging. Bobholz et al (78) coregistered MRI and hematoxylin-eosin–stained histologic images that were downsampled to the MRI resolution to compare the local texture information in both data sets. Comparison of the radiomic features from matching areas of the images identified a subset of metrics closely related between the two modalities. Although not directly revealing the biological underpinning of the metrics, these findings bring us closer to a solution as the biological meaning of hematoxylin-eosin stain patterns is significantly less ambiguous than are most MRI scans. In addition, knowing the close correlation between the low-resolution radiomic and pathomic features, the latter can then be related to the local phenotype with the help of pathologists. A step in this direction is described by McGarry et al (79,80), again using coregistered hematoxylin-eosin stain and multiparametric MRI data sets to develop local radiomic predictions of histologic parameters related to tumor grade in prostate cancer, although the unsupervised nature of the model does not reveal the direct contributions of different multiparametric MRI components to the predictions. Matched quantification of MRI and pathologic images was also used by Tomaszewski et al (81) for validation of the proposed feature of radiation therapy response. These examples illustrate the value in coregistration of in vivo and ex vivo images for detailed, spatially resolved insight into the relationships, shown pictorially in Figure 4. The local nature of the findings does not always translate directly to the main aim of radiomic analysis, which is focused on general per-patient signatures of survival and response. However, the local comparisons and observed correlation can shed light on the biological meaning of the radiomic metrics and can provide additional validation for observed signatures of outcome, thus enabling screening of features for relationships to histologic findings.
Figure 4:
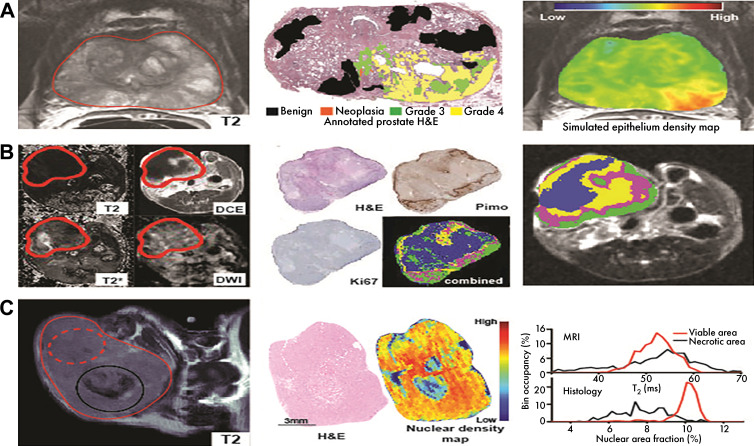
Coregistered histologic findings provide biological insight into image features. H&E = hematoxylin-eosin. A, McGarry et al (additional data provided by authors) developed model using multiparametric MRI information (left), trained on coregistered annotated hematoxylin-eosin–stained slides (middle) to model prostate epithelium density (right), relevant for tumor staging. B, Jardim-Perassi used multiparametric MRI (left), coregistered with histologic maps of viability, proliferation, and hypoxia (middle) to understand biological meaning of imaging habitats (right). DCE = dynamic contrast enhanced, DWI = diffusion-weighted imaging, Pimo = pimonidazole. C, Tomaszewski et al proposed T2-weighted MRI (left) histogram biomarker of radiation therapy response and used coregistered histologically derived nuclear density maps (middle) to demonstrate source of observed imaging changes through similarities in histogram features (right). Source.—References 80, 81, 90.
Habitat Imaging
Radiomic analysis has been widely applied in attempts to inform and quantify tumor heterogeneity. A separate approach explicitly aimed at identifying distinct tumor areas and cell subpopulations is represented by habitat imaging, which represents a middle ground between traditional whole-tumor and local per-voxel analysis as presented earlier. Combining images from multiple techniques, such as multiparametric MRI (82), or PET/CT (83), enables establishment of quantitative signatures used for delineation of distinct functional regions (habitats) within the tumor mass (84). The use of complex signatures from multidimensional information, an approach shared with radiomic analysis, leads to shared challenges in biological interpretation and validation of the findings. Relative habitat volumes have been reported as a predictor of survival (85) and genetic pathway dysregulation (86) using MRI or PET/CT data (87) for clustering. Limited insight into the biological meaning of the habitats can be provided by independent delineation of apparent tissue phenotypes such as necrosis and edema (86). With the help of careful histologic validation, Henning et al (88,89) tracked temporal dynamics of hypoxic, viable, and necrotic MRI habitats in a preclinical model of sarcoma following radiation therapy. Efforts to provide even more detailed biological insight are already available preclinically through per-pixel spatial coregistration of the images and corresponding histologic findings, thus demonstrating the use of MRI habitats for delineation of hypoxia, necrosis, and other conditions (90). The retrospective design of many clinical quantitative imaging studies prevents access to such insight. Prospective radiomic studies, including coregistered pathologic collection in the protocol, and early reports, such as the ovarian cancer case study (91) and continued efforts in renal carcinoma (92) or the Total Tumor Mapping trial in pancreatic cancer (ClinicalTrials.gov identifier: NCT03718650), will be necessary to shed light on the biological meaning of clinical image habitats.
Division of tumor and surrounding tissues into distinct physiologic subregions can also be used to focus the radiomic analysis to informative areas. Beig et al (93) performed feature extraction separately in edema, necrotic, and enhancing regions of brain tumors, showing their different informational content for progression-free survival prediction and association with gene expression. A similar approach was also used (94) to relate radiomic signatures to metabolic traits, as measured by MR spectroscopy. However, the increased number of features due to analysis of multiple regions requires larger cohort sizes and careful validation.
Biologically validated habitat imaging will be essential for its perhaps most promising clinical application—radiation therapy planning (95). As stressed by Enderling et al (96), knowledge of the spatial distribution of physiologic tumor subregions could allow for dose painting to optimize the response according to local radiosensitivity profiles. Early developments in these data-driven dose planning approaches based on image (radiomics) and dose shape (dosiomics) features are already promising for the reduction of radiation toxicity (97,98) and for enabling personalized dose prescription (99).
Discussion
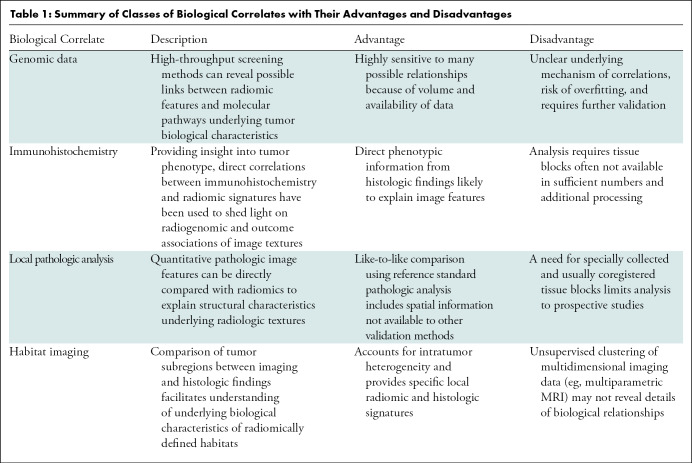
Our review showed significant recent effort in biological validation of radiomic findings. Four main classes of biological correlates and approaches used to date to inform the biological underpinnings of radiomics are identified, including gene expression data, protein expression from immunohistochemistry staining, microscopic histologic textures, and physiologic tumor habitats. The summary of these core classes and their evaluation is presented in Table 1.
Table 1:
Summary of Classes of Biological Correlates with Their Advantages and Disadvantages
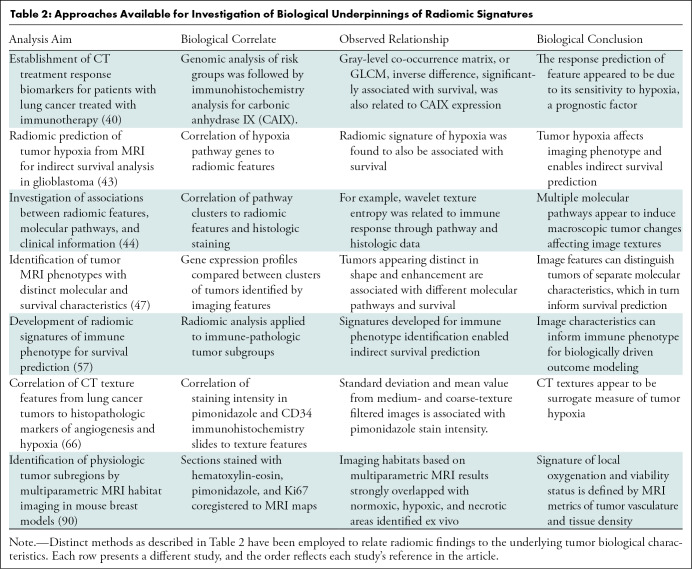
In a traditional radiomic pipeline, a signature of outcome, developed and validated in an independent training set, may be subsequently investigated for its association with a particular biological metric such as gene expression or IHC staining intensity. This method, as used among others by Sun et al (39) and Tunali et al (40), can strengthen the model, retrospectively informing the possible mechanism of outcome prediction. Conversely, the biological correlate can be used explicitly for model building, arriving at a radiomic signature indirectly associated with outcome because of the biological correlation, such as the negative prognostic value of tumor hypoxia, as in the studies by Beig et al (43). Although both approaches provide important validation and insight into the tumor biological characteristics, the more hypothesis-driven focus on the second method may deem it less biased for outcome analysis. Examples of the distinct methods designed to provide a biological insight into radiomic signatures, together with the findings and their significance, are presented in Table 2.
Table 2:
Approaches Available for Investigation of Biological Underpinnings of Radiomic Signatures
As the field develops, studies are increasingly expected to report at least one more correlation of the proposed radiomic model beyond the primary relationship with survival or other end point metrics. These can be incorporated at different stages of the process as a prescreening tool for feature selection or as the primary source of test data before survival calculation. The microscopic texture of histologic slides can also be used for additional insight.
Coclinical and preclinical experiments may also serve an important purpose in biological validation of radiomic findings. The controlled environment of animal studies enables the experimental interventions necessary to establish causal relationships between biology and radiomics and to offer precise, spatially coregistered histologic analysis for in-depth validation. However, the direct translatability of imaging findings between the spatial scales remains a challenge. Increasingly, clinical reports (eg, in the brain [78], prostate [80], or ovarian cancer [91]) demonstrate the feasibility and high value of three-dimensional printing–aided coregistration of in vivo and ex vivo images, providing spatially resolved insight into the biological meaning of local imaging characteristics.
Conclusions
To date, many radiomic studies present no validation of the findings beyond using an independent test cohort. This trend contributes to poor reproducibility and hence limited impact. With further recognition of the importance of biological understanding of the radiomic signatures, a standardized validation roadmap should be developed and deployed within the radiomics community. Many approaches, as summarized in our review, are already available for validation and biological context of the features. Going forward, we propose that reported studies should all strive to include such analysis, either as part of the model building or subsequent validation, to provide a hypothesis for the biological mechanism of the observed relationship. This will enable the discussion of the biological characteristics underlying the findings to become a standard in the field and enforced in the peer-review process.
Supported by National Institutes of Health grants U01 CA143062 and U54 CA143970.
Disclosures of Conflicts of Interest: M.R.T. disclosed no relevant relationships. R.J.G. Activities related to the present article: institution received grant from National Cancer Institute. Activities not related to the present article: holds stock/stock options in HealthMyne; has research sponsored in kind with software access by SRA. Other relationships: has patents issued for intellectual property.
Abbreviation:
- IHC
- immunohistochemistry
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48(4):441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016;6(11):e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14(12):749–762. [DOI] [PubMed] [Google Scholar]
- 4.Zwanenburg A, Leger S, Vallières M, Löck S. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallières M, Zwanenburg A, Badic B, Cheze Le Rest C, Visvikis D, Hatt M. Responsible Radiomics Research for Faster Clinical Translation. J Nucl Med 2018;59(2):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Z, Traverso A, van Soest J, Dekker A, Wee L. Technical Note: Ontology-guided radiomics analysis workflow (O-RAW). Med Phys 2019;46(12):5677–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Z, Zhovannik I, Traverso A, et al. Distributed radiomics as a signature validation study using the Personal Health Train infrastructure. Sci Data 2019;6(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin DL, Rodriguez C, Shah P, Beaulieu C. iPad: Semantic annotation and markup of radiological images. AMIA Annu Symp Proc 2008;2008:626–630. [PMC free article] [PubMed] [Google Scholar]
- 11.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25(6):675–680. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Kim J, Qu F, et al. CT Features Associated with Epidermal Growth Factor Receptor Mutation Status in Patients with Lung Adenocarcinoma. Radiology 2016;280(1):271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kundu S, Itkin M, Gervais DA, et al. The IR Radlex Project: an interventional radiology lexicon--a collaborative project of the Radiological Society of North America and the Society of Interventional Radiology. J Vasc Interv Radiol 2009;20(7 Suppl):S275–S277. [DOI] [PubMed] [Google Scholar]
- 14.Yip SSF, Liu Y, Parmar C, et al. Associations between radiologist-defined semantic and automatically computed radiomic features in non-small cell lung cancer. Sci Rep 2017;7(1):3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tunali I, Stringfield O, Guvenis A, et al. Radial gradient and radial deviation radiomic features from pre-surgical CT scans are associated with survival among lung adenocarcinoma patients. Oncotarget 2017;8(56):96013–96026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gevaert O, Echegaray S, Khuong A, et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep 2017;7(1):41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koay EJ, Lee Y, Cristini V, et al. A Visually Apparent and Quantifiable CT Imaging Feature Identifies Biophysical Subtypes of Pancreatic Ductal Adenocarcinoma. Clin Cancer Res 2018;24(23):5883–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amer AM, Zaid M, Chaudhury B, et al. Imaging-based biomarkers: Changes in the tumor interface of pancreatic ductal adenocarcinoma on computed tomography scans indicate response to cytotoxic therapy. Cancer 2018;124(8):1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012;3(6):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor JP, Rose CJ, Jackson A, et al. DCE-MRI biomarkers of tumour heterogeneity predict CRC liver metastasis shrinkage following bevacizumab and FOLFOX-6. Br J Cancer 2011;105(1):139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alic L, van Vliet M, van Dijke CF, Eggermont AM, Veenland JF, Niessen WJ. Heterogeneity in DCE-MRI parametric maps: a biomarker for treatment response? Phys Med Biol 2011;56(6):1601–1616. [DOI] [PubMed] [Google Scholar]
- 22.Eary JF, O’Sullivan F, O’Sullivan J, Conrad EU. Spatial heterogeneity in sarcoma 18F-FDG uptake as a predictor of patient outcome. J Nucl Med 2008;49(12):1973–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skogen K, Ganeshan B, Good C, Critchley G, Miles K. Measurements of heterogeneity in gliomas on computed tomography relationship to tumour grade. J Neurooncol 2013;111(2):213–219. [DOI] [PubMed] [Google Scholar]
- 24.Rees JH, Smirniotopoulos JG, Jones RV, Wong K. Glioblastoma multiforme: radiologic-pathologic correlation. RadioGraphics 1996;16(6):1413–1438; quiz 1462–1463. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Liu S, Qu F, Li Q, Cheng R, Ye Z. Tumor heterogeneity assessed by texture analysis on contrast-enhanced CT in lung adenocarcinoma: association with pathologic grade. Oncotarget 2017;8(32):53664–53674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Li H, Guo W, et al. Deciphering Genomic Underpinnings of Quantitative MRI-based Radiomic Phenotypes of Invasive Breast Carcinoma. Sci Rep 2015;5(1):17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graim K, Liu TT, Achrol AS, et al. Revealing cancer subtypes with higher-order correlations applied to imaging and omics data. BMC Med Genomics 2017;10(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rios Velazquez E, Parmar C, Liu Y, et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res 2017;77(14):3922–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumor phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5(1):4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012;264(2):387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giaccone G, Herbst RS, Manegold C, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 1. J Clin Oncol 2004;22(5):777–784. [DOI] [PubMed] [Google Scholar]
- 34.Masui H, Kawamoto T, Sato JD, Wolf B, Sato G, Mendelsohn J. Growth inhibition of human tumor cells in athymic mice by anti-epidermal growth factor receptor monoclonal antibodies. Cancer Res 1984;44(3):1002–1007. [PubMed] [Google Scholar]
- 35.Yip SS, Kim J, Coroller TP, et al. Associations Between Somatic Mutations and Metabolic Imaging Phenotypes in Non-Small Cell Lung Cancer. J Nucl Med 2017;58(4):569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CT, Yen RF, Cheng MF, et al. Correlation of F-18 fluorodeoxyglucose-positron emission tomography maximal standardized uptake value and EGFR mutations in advanced lung adenocarcinoma. Med Oncol 2010;27(1):9–15. [DOI] [PubMed] [Google Scholar]
- 37.Henriksson E, Kjellen E, Wahlberg P, Ohlsson T, Wennerberg J, Brun E. 2-Deoxy-2-[18F] fluoro-D-glucose uptake and correlation to intratumoral heterogeneity. Anticancer Res 2007;27(4B):2155–2159. [PubMed] [Google Scholar]
- 38.Lee SM, Bae SK, Jung SJ, Kim CK. FDG uptake in non-small cell lung cancer is not an independent predictor of EGFR or KRAS mutation status: a retrospective analysis of 206 patients. Clin Nucl Med 2015;40(12):950–958. [DOI] [PubMed] [Google Scholar]
- 39.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19(9):1180–1191. [DOI] [PubMed] [Google Scholar]
- 40.Tunali I, Tan Y, Gray JE, et al. Hypoxia-related radiomics predict immunotherapy response: A multi-cohort study of NSCLC. bioRxiv. Posted April 4, 2020. [Google Scholar]
- 41.Mu W, Jiang L, Shi Y, et al. Prediction of immunotherapy response using deep learning of PET/CT images. Can Res 2020 (in revision). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu W, Jiang L, Zhang J, et al. Radiomics based treatment decision support for NSCLC. Nat Commun 2020 (in revision). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beig N, Patel J, Prasanna P, et al. Radiogenomic analysis of hypoxia pathway is predictive of overall survival in Glioblastoma. Sci Rep 2018;8(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. eLife 2017;6:e23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J, Li B, Sun X, et al. Heterogeneous Enhancement Patterns of Tumor-adjacent Parenchyma at MR Imaging Are Associated with Dysregulated Signaling Pathways and Poor Survival in Breast Cancer. Radiology 2017;285(2):401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, Cui Y, Sun X, et al. Unsupervised Clustering of Quantitative Image Phenotypes Reveals Breast Cancer Subtypes with Distinct Prognoses and Molecular Pathways. Clin Cancer Res 2017;23(13):3334–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015;7(303):303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu TT, Achrol AS, Mitchell LA, et al. Magnetic resonance perfusion image features uncover an angiogenic subgroup of glioblastoma patients with poor survival and better response to antiangiogenic treatment. Neuro Oncol 2017;19(7):997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panth KM, Leijenaar RT, Carvalho S, et al. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol 2015;116(3):462–466. [DOI] [PubMed] [Google Scholar]
- 50.Zinn PO, Singh SK, Kotrotsou A, et al. A Coclinical Radiogenomic Validation Study: Conserved Magnetic Resonance Radiomic Appearance of Periostin-Expressing Glioblastoma in Patients and Xenograft Models. Clin Cancer Res 2018;24(24):6288–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu W, Parmar C, Grossmann P, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol 2016;6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SE, Han K, Kwak JY, Lee E, Kim EK. Radiomics of US texture features in differential diagnosis between triple-negative breast cancer and fibroadenoma. Sci Rep 2018;8(1):13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanduleanu S, Woodruff HC, de Jong EEC, et al. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radiother Oncol 2018;127(3):349–360. [DOI] [PubMed] [Google Scholar]
- 54.Yip SS, Coroller TP, Sanford NN, Mamon H, Aerts HJ, Berbeco RI. Relationship between the Temporal Changes in Positron-Emission-Tomography-Imaging-Based Textural Features and Pathologic Response and Survival in Esophageal Cancer Patients. Front Oncol 2016;6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z, Zhang XY, Shi YJ, et al. Radiomics Analysis for Evaluation of Pathological Complete Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer. Clin Cancer Res 2017;23(23):7253–7262. [DOI] [PubMed] [Google Scholar]
- 56.Yin Q, Hung SC, Wang L, et al. Associations between Tumor Vascularity, Vascular Endothelial Growth Factor Expression and PET/MRI Radiomic Signatures in Primary Clear-Cell-Renal-Cell-Carcinoma: Proof-of-Concept Study. Sci Rep 2017;7(1):43356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang C, Hobbs B, Amer A, et al. Development of an Immune-Pathology Informed Radiomics Model for Non-Small Cell Lung Cancer. Sci Rep 2018;8(1):1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ha S, Park S, Bang JI, Kim EK, Lee HY. Metabolic Radiomics for Pretreatment 18F-FDG PET/CT to Characterize Locally Advanced Breast Cancer: Histopathologic Characteristics, Response to Neoadjuvant Chemotherapy, and Prognosis. Sci Rep 2017;7(1):1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gillies RJ, Schornack PA, Secomb TW, Raghunand N. Causes and effects of heterogeneous perfusion in tumors. Neoplasia 1999;1(3):197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammond EM, Asselin MC, Forster D, O’Connor JPB, Senra JM, Williams KJ. The meaning, measurement and modification of hypoxia in the laboratory and the clinic. Clin Oncol (R Coll Radiol) 2014;26(5):277–288. [DOI] [PubMed] [Google Scholar]
- 61.Tatum JL, Kelloff GJ, Gillies RJ, et al. Hypoxia: importance in tumor biology, noninvasive measurement by imaging, and value of its measurement in the management of cancer therapy. Int J Radiat Biol 2006;82(10):699–757. [DOI] [PubMed] [Google Scholar]
- 62.Krohn KA, Link JM, Mason RP. Molecular imaging of hypoxia. J Nucl Med 2008;49(Suppl 2):129S–148S. [DOI] [PubMed] [Google Scholar]
- 63.Peeters SG, Zegers CM, Yaromina A, Van Elmpt W, Dubois L, Lambin P. Current preclinical and clinical applications of hypoxia PET imaging using 2-nitroimidazoles. Q J Nucl Med Mol Imaging 2015;59(1):39–57. [PubMed] [Google Scholar]
- 64.Sanduleanu S, Wiel AMAV, Lieverse RIY, et al. Hypoxia PET Imaging with [18F]-HX4-A Promising Next-Generation Tracer. Cancers (Basel) 2020;12(5):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ragnum HB, Vlatkovic L, Lie AK, et al. The tumour hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br J Cancer 2015;112(2):382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 2013;266(1):326–336. [DOI] [PubMed] [Google Scholar]
- 67.Muzi M, Wolsztynski E, Fink JR, et al. Assessment of the Prognostic Value of Radiomic Features in 18F-FMISO PET Imaging of Hypoxia in Postsurgery Brain Cancer Patients: Secondary Analysis of Imaging Data from a Single-Center Study and the Multicenter ACRIN 6684 Trial. Tomography 2020;6(1):14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sörensen A, Carles M, Bunea H, et al. Textural features of hypoxia PET predict survival in head and neck cancer during chemoradiotherapy. Eur J Nucl Med Mol Imaging 2020;47(5):1056–1064. [DOI] [PubMed] [Google Scholar]
- 69.Crispin-Ortuzar M, Apte A, Grkovski M, et al. Predicting hypoxia status using a combination of contrast-enhanced computed tomography and [18F]-Fluorodeoxyglucose positron emission tomography radiomics features. Radiother Oncol 2018;127(1):36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Jong EEC, van Elmpt W, Leijenaar RTH, et al. [18F]FDG PET/CT-based response assessment of stage IV non-small cell lung cancer treated with paclitaxel-carboplatin-bevacizumab with or without nitroglycerin patches. Eur J Nucl Med Mol Imaging 2017;44(1):8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta R, Kurc T, Sharma A, Almeida JS, Saltz J. The Emergence of Pathomics. Curr Pathobiol Rep 2019;7(3):73–84. [Google Scholar]
- 72.Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A 2018;115(13):E2970–E2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011;3(108):108ra113. [DOI] [PubMed] [Google Scholar]
- 74.Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7(1):12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saltz J, Almeida J, Gao Y, et al. Towards Generation, Management, and Exploration of Combined Radiomics and Pathomics Datasets for Cancer Research. AMIA Jt Summits Transl Sci Proc 2017;2017:85–94. [PMC free article] [PubMed] [Google Scholar]
- 76.Chaddad A, Daniel P, Niazi T. Radiomics Evaluation of Histological Heterogeneity Using Multiscale Textures Derived From 3D Wavelet Transformation of Multispectral Images. Front Oncol 2018;8:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geady C, Keller H, Siddiqui I, Bilkey J, Dhani NC, Jaffray DA. Bridging the gap between micro- and macro-scales in medical imaging with textural analysis - A biological basis for CT radiomics classifiers? Phys Med 2020;72:142–151. [DOI] [PubMed] [Google Scholar]
- 78.Bobholz SA, Lowman AK, Barrington A, et al. Radiomic Features of Multiparametric MRI Present Stable Associations With Analogous Histological Features in Patients With Brain Cancer. Tomography 2020;6(2):160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McGarry SD, Bukowy JD, Iczkowski KA, et al. Gleason Probability Maps: A Radiomics Tool for Mapping Prostate Cancer Likelihood in MRI Space. Tomography 2019;5(1):127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGarry SD, Hurrell SL, Iczkowski KA, et al. Radio-pathomic Maps of Epithelium and Lumen Density Predict the Location of High-Grade Prostate Cancer. Int J Radiat Oncol Biol Phys 2018;101(5):1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tomaszewski MR, Dominguez-Viqueira W, Ortiz A, et al. Heterogeneity analysis of MRI T2 maps for measurement of early tumor response to radiotherapy. NMR in Biomedicine 2020;e4454. [DOI] [PubMed] [Google Scholar]
- 82.Sun Y, Reynolds HM, Parameswaran B, et al. Multiparametric MRI and radiomics in prostate cancer: a review. Australas Phys Eng Sci Med 2019;42(1):3–25. [DOI] [PubMed] [Google Scholar]
- 83.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging 2013;40(1):133–140. [DOI] [PubMed] [Google Scholar]
- 84.Napel S, Mu W, Jardim-Perassi BV, Aerts HJWL, Gillies RJ. Quantitative imaging of cancer in the postgenomic era: Radio(geno)mics, deep learning, and habitats. Cancer 2018;124(24):4633–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stringfield O, Arrington JA, Johnston SK, et al. Multiparameter MRI Predictors of Long-Term Survival in Glioblastoma Multiforme. Tomography 2019;5(1):135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dextraze K, Saha A, Kim D, et al. Spatial habitats from multiparametric MR imaging are associated with signaling pathway activities and survival in glioblastoma. Oncotarget 2017;8(68):112992–113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu J, Gensheimer MF, Zhang N, et al. Tumor Subregion Evolution-Based Imaging Features to Assess Early Response and Predict Prognosis in Oropharyngeal Cancer. J Nucl Med 2020;61(3):327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Henning EC, Azuma C, Sotak CH, Helmer KG. Multispectral quantification of tissue types in a RIF-1 tumor model with histological validation. Part I. Magn Reson Med 2007;57(3):501–512. [DOI] [PubMed] [Google Scholar]
- 89.Henning EC, Azuma C, Sotak CH, Helmer KG. Multispectral tissue characterization in a RIF-1 tumor model: monitoring the ADC and T2 responses to single-dose radiotherapy. Part II. Magn Reson Med 2007;57(3):513–519. [DOI] [PubMed] [Google Scholar]
- 90.Jardim-Perassi BV, Huang S, Dominguez-Viqueira W, et al. Multiparametric MRI and Coregistered Histology Identify Tumor Habitats in Breast Cancer Mouse Models. Cancer Res 2019;79(15):3952–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vargas HA, Huang EP, Lakhman Y, et al. Radiogenomics of High-Grade Serous Ovarian Cancer: Multireader Multi-Institutional Study from the Cancer Genome Atlas Ovarian Cancer Imaging Research Group. Radiology 2017;285(2):482–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crispin-Ortuzar M, Gehrung M, Ursprung S, et al. 3D-printed moulds of renal tumours for image-guided tissue sampling in the clinical setting. bioRxiv. Posted March 4, 2020. [Google Scholar]
- 93.Beig N, Bera K, Prasanna P, et al. Radiogenomic-Based Survival Risk Stratification of Tumor Habitat on Gd-T1w MRI Is Associated with Biological Processes in Glioblastoma. Clin Cancer Res 2020;26(8):1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lopez CJ, Nagornaya N, Parra NA, et al. Association of Radiomics and Metabolic Tumor Volumes in Radiation Treatment of Glioblastoma Multiforme. Int J Radiat Oncol Biol Phys 2017;97(3):586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scalco E, Rizzo G. Texture analysis of medical images for radiotherapy applications. Br J Radiol 2017;90(1070):20160642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Enderling H, Alfonso JCL, Moros E, Caudell JJ, Harrison LB. Integrating Mathematical Modeling into the Roadmap for Personalized Adaptive Radiation Therapy. Trends Cancer 2019;5(8):467–474. [DOI] [PubMed] [Google Scholar]
- 97.Gabryś HS, Buettner F, Sterzing F, Hauswald H, Bangert M. Design and Selection of Machine Learning Methods Using Radiomics and Dosiomics for Normal Tissue Complication Probability Modeling of Xerostomia. Front Oncol 2018;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rossi L, Bijman R, Schillemans W, et al. Texture analysis of 3D dose distributions for predictive modelling of toxicity rates in radiotherapy. Radiother Oncol 2018;129(3):548–553. [DOI] [PubMed] [Google Scholar]
- 99.Kjellsson Lindblom E, Ureba A, Dasu A, et al. Impact of SBRT fractionation in hypoxia dose painting - Accounting for heterogeneous and dynamic tumor oxygenation. Med Phys 2019;46(5):2512–2521. [DOI] [PubMed] [Google Scholar]