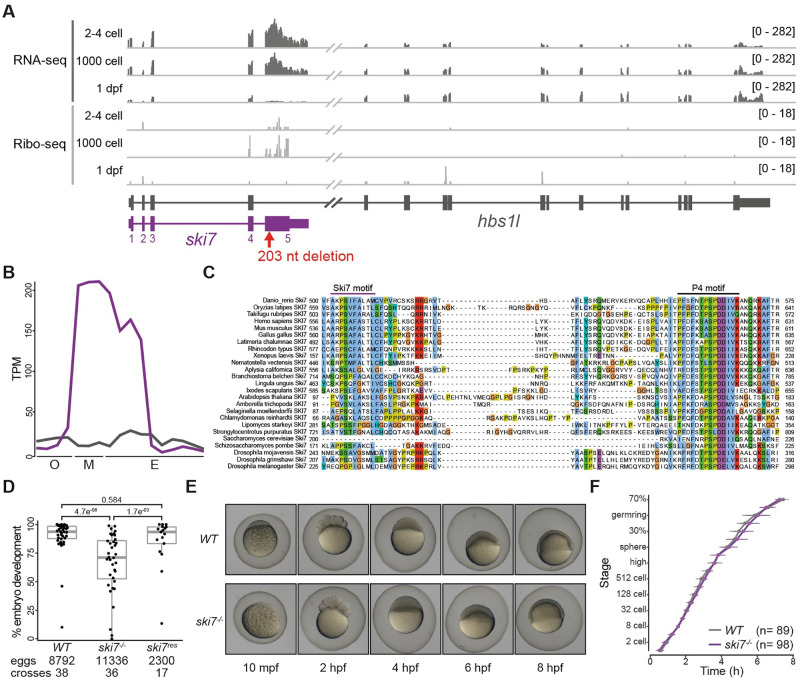
Fig 1. Ski7 mutant fish produce reduced numbers of developing offspring.
(A) Expression profile of the hbs1l (gray)/ski7 (purple) locus during embryogenesis. Ski7 is highly expressed in the early embryo. RNA-seq is shown in dark gray and Ribo-seq in light gray; numbers in brackets indicate expression levels; the red arrow indicates the site of the ski7 mutation. (B) Ski7 mRNA peaks during the oocyte-to-embryo transition. Transcript-specific quantification of ski7 (purple) and hbs1l (gray) RNA from polyA+ RNA-seq during oogenesis (O: oocyte stages I-IV), in mature eggs (M: unactivated, activated, fertilized) and during embryogenesis (E: 2–4 cell, 256 cell, 1000 cell, oblong, dome, shield, bud, 1 dpf, 2 dpf, 5 dpf [34]); TPM, transcripts per million. (C) Ski7 protein is highly conserved. Protein sequence alignment of the conserved C-terminus of Ski7 across different organisms. The two most conserved motifs, the Ski7-like motif and the Patch (P4)-like motif, are highlighted. (D) Ski7-/- fish produce fewer embryos that develop. Number of embryos that progress beyond the one-cell stage of wild-type (WT), ski7-/- and ski7res (ski7-/-; tg[actb2:ski7-GFP]) fish. Transgenic ski7-GFP rescues the decreased number of viable progeny observed in ski7-/- fish. P-adjusted values from Kruskal-Wallis with Dunn’s multiple comparison test. (E) Ski7-/- embryos develop normally. Representative images of WT and ski7-/- embryos from 10 minutes post-fertilisation (mpf) to 8 hours post-fertilisation (hpf). (F) Quantification of embryo development in WT and ski7-/- embryos. Ski7-/- embryos that undergo cell cleavage develop at normal rate.