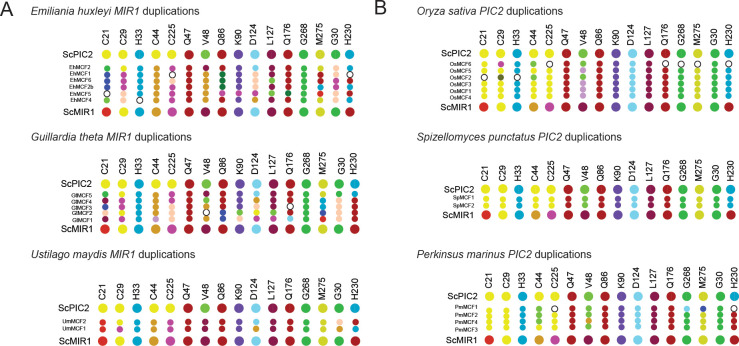
Figure 5. Conservation of selected residues in the PIC2/MIR1 family of transporters.
The tree topology is identical to that shown in Figure 2. Amino acids are colored according to the key, and insertion/deletion events that lead to gaps within the alignment are indicated by the hollow circles. P indicates position of S. cerevisiae PIC2, and M indicates S. cerevisiae MIR1. Small dots indicate that the residue is identical to that of PIC2 (shown at the top), and large dots indicate differences.

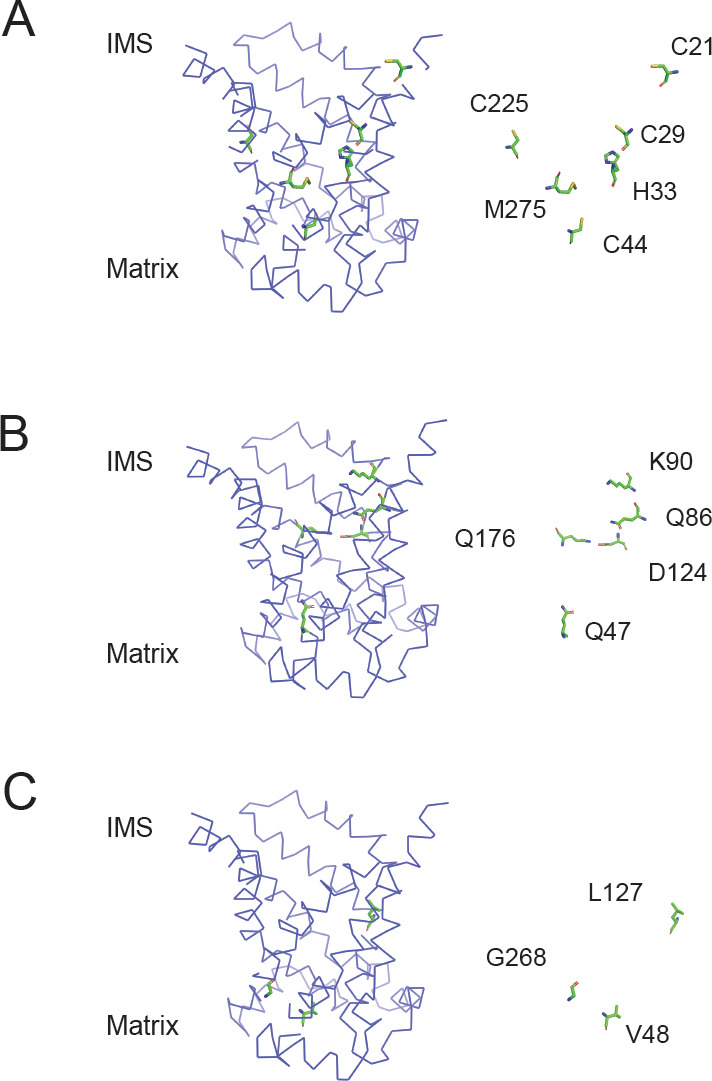
Figure 5—figure supplement 1. Map of the residues found in gene duplications.
Figure 5—figure supplement 2. Position of residues mutated in the aqueous binding pocket of PIC2.