Figure 6. Expression of PIC2 and variants in L. lactis.
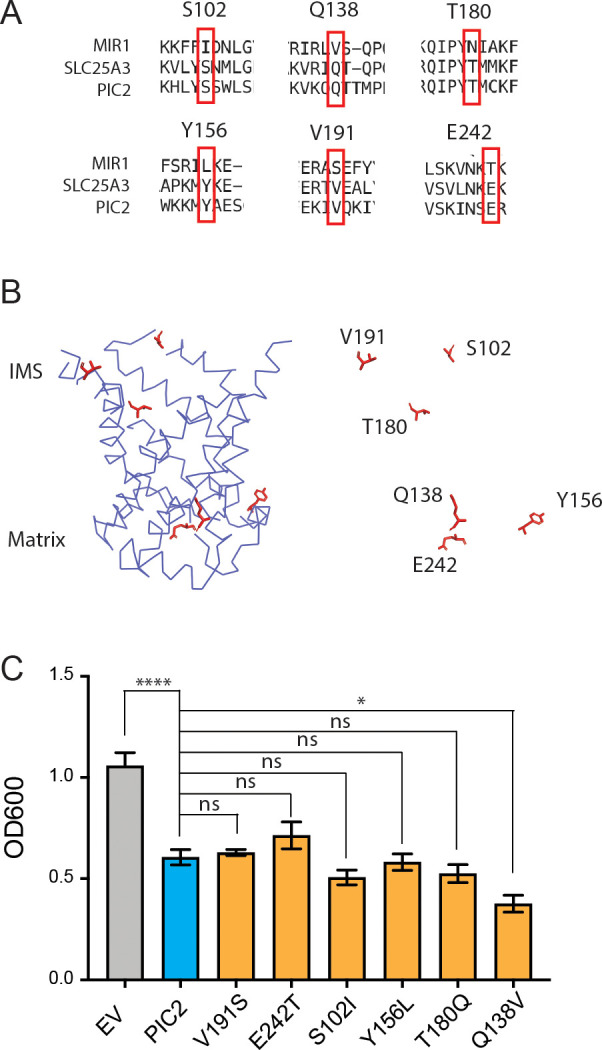
(A) Immunoblot of L. lactis extracts expressing empty vector (EV), wild-type PIC2 (WT), or a given PIC2 variant in which each of the listed residues was converted to an alanine prepared from equal numbers of cells based on optical density after induction with nisin. (B) Growth of L. lactis expressing EV, wild-type PIC2 (WT), or a given PIC2 variant in Ag+-containing media. Each bar represents the median of 10–18 independent cultures with 95% confidence interval as error bars (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 based on one-way ANOVA relative to PIC2 wild-type control). The color of the bar indicates one of four major groupings: Cu-binding (orange), structural motifs or contact points (gray), evolutionarily conserved and present in the aqueous binding pocket of the transporter (green), and unstable in L. lactis (yellow). (C) As described in (B) except L. lactis strains were grown in AsO43−-containing media.
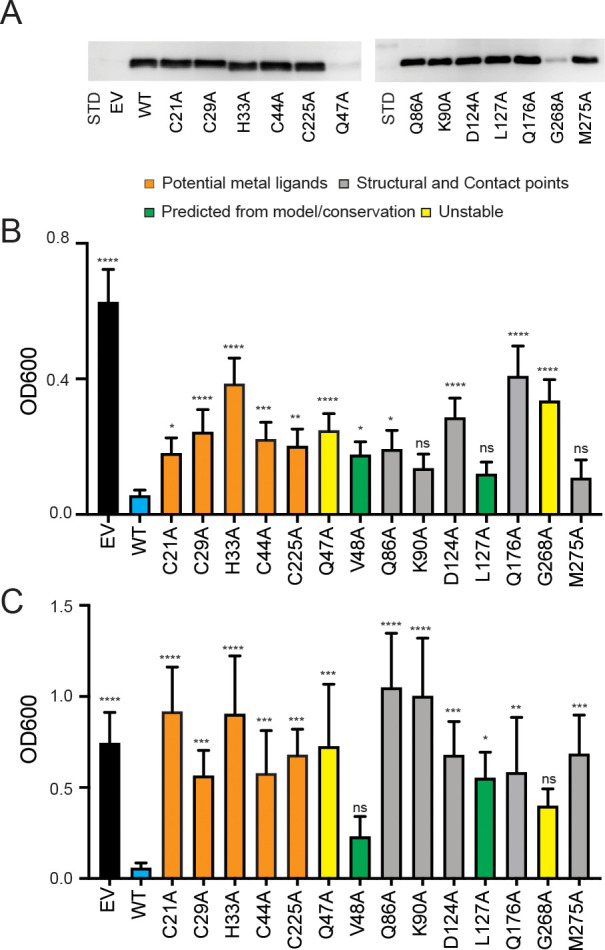
Figure 6—figure supplement 1. Protein expression in L. lactis.
Figure 6—figure supplement 2. Immunoblot analysis of PIC2 expressed in L. lactis.

Figure 6—figure supplement 3. Substitution of PIC2 residues for MIR1 residues.