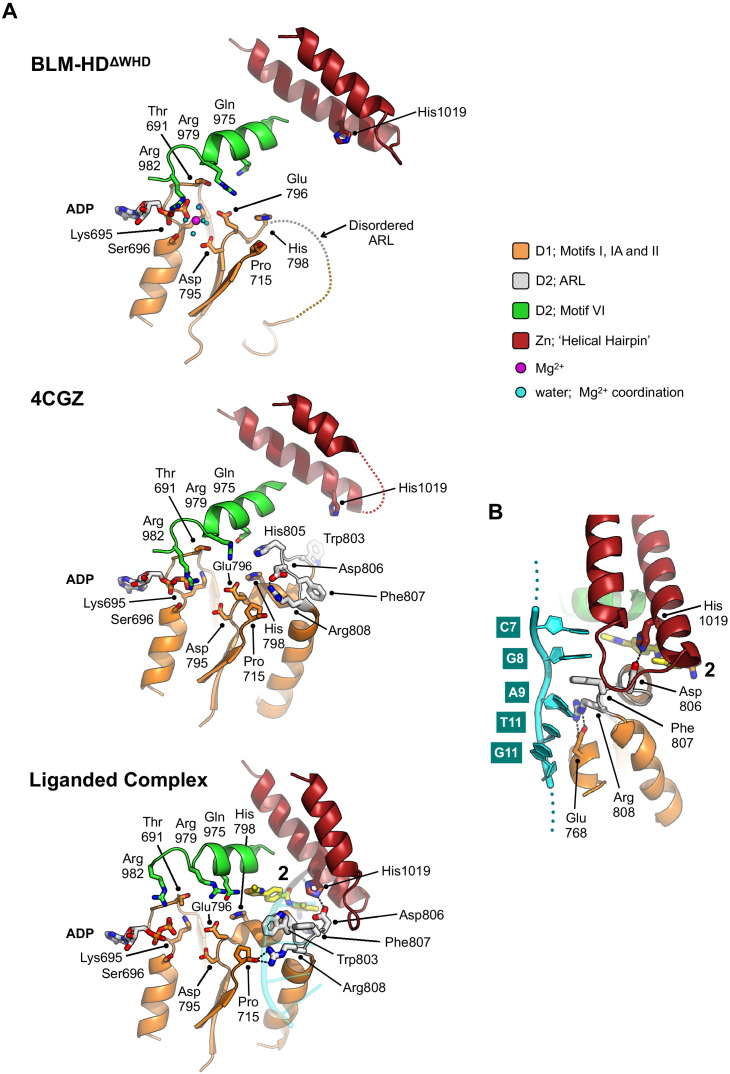
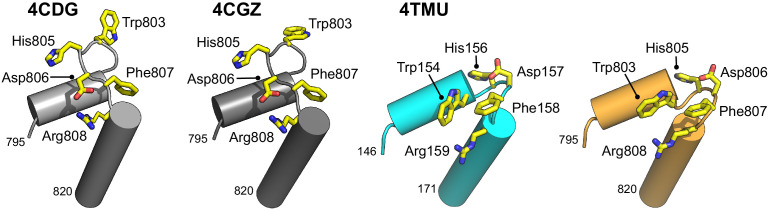
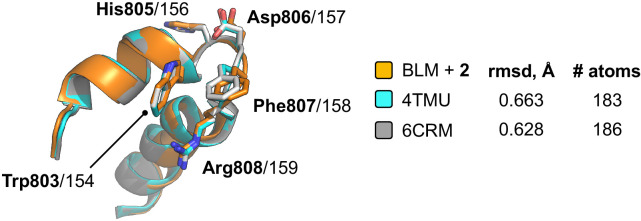
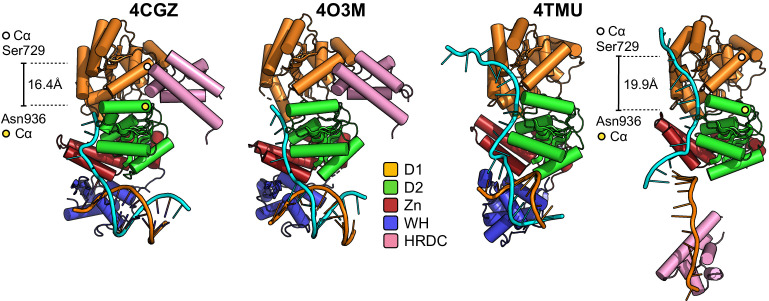
Figure 4. Structural transitions around the aromatic rich loop.
(A) Molecular secondary structure cartoons for the region surrounding the aromatic rich loop (ARL) of BLM-HDΔWHD (top), PDB entry 4CGZ; BLM-HD in complex with DNA (middle) and liganded complex; BLM-HDΔWHD in complex with ADP, ssDNA-15mer and 2 (bottom). The side chains for key amino acid residues are shown in stick representation, with carbon atoms coloured according to their respective domains (see associated key). Bound ADP and 2 are also shown in stick representation, with carbon atoms coloured grey and yellow, respectively. (B) Expanded and rotated view highlighting the interactions made between the ARL and ssDNA-15mer oligonucleotide (cartoon coloured cyan) in the liganded complex, also showing the relative position of compound 2. Potential hydrogens bonds are represented by black dotted lines.