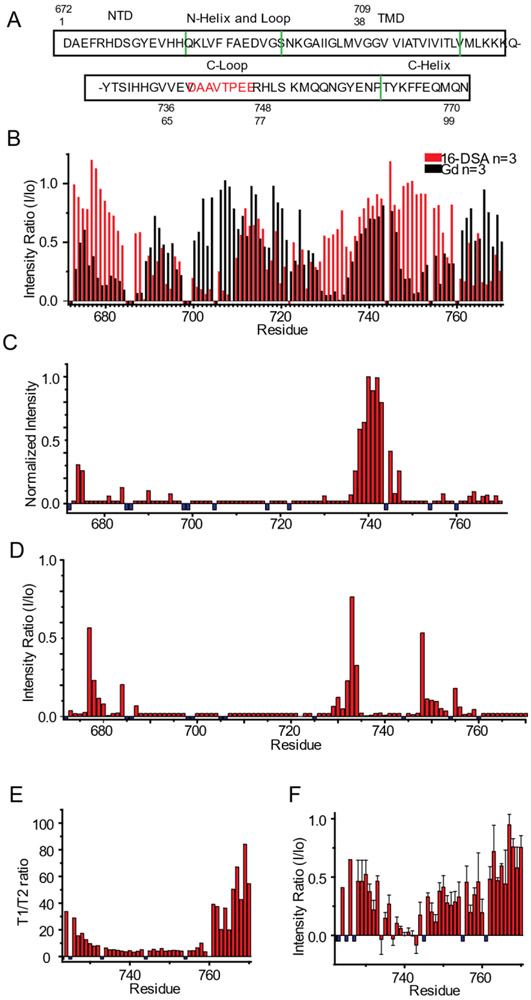
Figure 6. Use of NMR to probe the topology and dynamics of C99 in SCOR bicelles.
A) Sequence of human C99 with numbering based either on C99 only (sites 1-99) or on the parent full-length amyloid precursor protein (672-770). Green lines indicate boundaries of secondary structural elements as determined in this work from analysis of the backbone NMR resonance chemical shifts and paramagnetic data. Amino acids in the C-loop previously identified as being involved in transient amide backbone hydrogen bonding are identified in red.49 B) Topology of C99 in SCOR bicelles as mapped by site-specific TROSY NMR peak linebroadening due to site access by lipophilic (16-DSA) or aqueous (Gd-DTPA) paramagnetic probes. Mean intensity ratios (probe-exposed versus control peak height) are presented for 16-DSA (red) and Gd-DTPA (black). The U-15N-C99 concentrations for all experiments were between ~200 and 400 μM in 5-10 wt% SCOR bicelles, q = 0.33, in NMR buffer. Error bars for these n=3 data are not shown here (to avoid clutter), but are displayed in Figure S6. C) Effect on C99 peak intensities of adding both excess 16-DSA (to 12 mol %), and Gd-DTPA (to 3mM) to a 300μM U-15N-C99 sample in 7 wt % SCOR bicelles and NMR buffer at 45 °C. n=1 D) CLEANEX-PM amide exchange analysis of peak height ratios for a 50 ms mixing time experiment. Ratios are of 50 ms mixing time versus no mixing time. E) NMR 15N T1/T2 relaxation time ratios for the cytosolic domain of C99. T1 and T2 analyses are derived from a 300 μM U-15N-C99 sample in 5 wt % SCOR bicelles q=0.33 in NMR buffer at 45 °C. n=1 F) Intensity ratios for 1H-15N NOE signals from a 300 μM C99 sample in 10 wt % SCOR bicelles, q=0.33, in NMR buffer at 45 °C. n=2 Small negative values in B are residues which were not assigned or were too broad; for C-F, these values are represented by small negative blue bars.