Abstract
Lytic polysaccharide monooxygenases (LPMOs) are mononuclear copper enzymes that catalyse the oxidative cleavage of glycosidic bonds. They are characterised by two histidine residues that coordinate copper in a configuration termed the Cu-histidine brace. Although first identified in bacteria and fungi, LPMOs have since been found in all biological kingdoms. LPMOs are now included in commercial enzyme cocktails used in industrial biorefineries. This has led to increased process yield due to the synergistic action of LPMOs with glycoside hydrolases. However, the introduction of LPMOs makes control of the enzymatic step in industrial stirred-tank reactors more challenging, and the operational stability of the enzymes is reduced. It is clear that much is still to be learned about the interaction between LPMOs and their complex natural and industrial environments, and fundamental scientific studies are required towards this end. Several atomic-resolution structures have been solved providing detailed information on the Cu-coordination sphere and the interaction with the polysaccharide substrate. However, the molecular mechanisms of LPMOs are still the subject of intense investigation; the key question being how the proteinaceous environment controls the copper cofactor towards the activation of the O-O bond in O2 and cleavage of the glycosidic bonds in polysaccharides. The need for biochemical characterisation of each putative LPMO is discussed based on recent reports showing that not all proteins with a Cu-histidine brace are enzymes.
Keywords: bioethanol, copper, lignocellulose, LPMO
Introduction
The replacement of liquid fossil transportation fuels with ethanol produced from agricultural and forest lignocellulosic residues is now technically possible [1]. However, the first commercial lignocellulosic biorefineries have suffered competition from historically low crude oil prices, which has made them economically strained. Although the biotechnological aspects of the process are in place, further improvements are required in process economy. Efficient saccharification takes place during incubation of the lignocellulosic material with the enzyme cocktail, at a slightly elevated temperature (typically 50°C) and constant pH (typically around pH 5), while the mixture is stirred. Enzyme cocktails consisting of many different types of enzymes are required for saccharification of the complex polysaccharide components of lignocellulose. When bioethanol is the desired product, yeast is added after 3–5 days of incubation to ferment the released glucose and xylose to ethanol. The residual material is high in lignin, and can be incinerated to produce steam and electricity [1]. Alternatively, it could potentially be used to produce marine fuel [2].
Lignocellulose is a complex recalcitrant matrix dominated by lignin, cellulose and hemicellulose, which provides the plant tissue with strength and durability. The most abundant polysaccharide in lignocellulose is cellulose, and several enzymes are involved in its decomposition into glucose. The process requires the synergistic action of endo-acting glycosyl hydrolases (endoglucanase), processive exo-hydrolases acting from the reducing end (cellobiohydrolase I), and processive exo-hydrolases acting from the non-reducing end (cellobiohydrolase II) of the cellulose chains. The overall reaction is driven towards completion by β-glycosidases through the alleviation of cellobiose product inhibition of the exo-enzymes [3–5]. Despite early reports indicating that an oxidative enzymatic step was crucial for cellulose saccharification [6–8], this notion was not fully accepted until the discovery of lytic polysaccharide monooxygenases (LPMOs) [9,10]. LPMOs initiate the saccharification of cellulose by oxidative cleavage of internal glucosidic bonds that are not accessible to the endo-acting hydrolases. LPMOs are mononuclear copper enzymes that are particularly common in the secretomes of saprophytic fungi feeding on lignocellulosic materials such as wood. This review discusses the Cu-active site of LPMOs, and how protein structure together with oxygen activation contribute to a catalytic cleavage.
LPMOs are classified into several enzyme families
LPMOs are classified in the Carbohydrate-Active EnZymes database (CAZy). They are grouped in enzyme families termed Auxiliary Activities (AAs) that catalogue the redox enzymes involved in carbohydrate degradation [11]. LPMOs have been demonstrated to have substrate competency towards a diverse group of polysaccharides, for example starch, xylan, xyloglucans and other hemicelluloses [12–16]. However, the two most studied activities are those on cellulose and chitin. Based on sequence similarity LPMOs are currently classified as AA9–AA11 and AA13–AA16 in the CAZy database. The most well characterised LPMOs are from the families AA9 and AA10, with multiple examples of biochemical characterisations and entries in the Protein Data Bank (PDB). While the first LPMOs were identified through fractionation of fungal secretomes in combination with saccharification assays [17], newer LPMOs have often been identified by data mining of genomic sequences. AA14 has recently been defined as a LPMO family, based on the characterisation of two enzymes from the white-rot basidiomycete Pycnoporus coccineus (now Trametes coccineus) [18]. The two T. coccineus enzymes demonstrated no enzymatic activity when used alone on 11 different polysaccharides, however, when incubated with either different Trichoderma reesei enzyme cocktails or with a GH11 xylanase, a synergistic boost was observed in the saccharification of xylan bound to cellulose in woody biomasses. Since no LPMO activity could be demonstrated on isolated polysaccharides, more detailed biochemical characterisation of the two enzymes is needed to deduce their enzymatic mechanisms. Interestingly, the presence of LPMOs in nature has recently expanded outside microorganisms, with examples being found in insects, animals and ferns [19,20]. The AA15 family has been discovered in the gut of the insect Thermobia domestica [20]. Biochemically characterised members of the AA15 family were found to degrade both cellulose and chitin. The most recent addition is the AA16 family, where an LPMO from the ascomycete Aspergillus aculeatus was heterologously expressed and biochemically shown to oxidatively cleave cellulose at the C1 position [21]. However, the amount of product formed was very low, and more thorough biochemical characterisation and its crystal structure are required to elucidate the enzymatic nature of this LPMO family.
In 2016, a protein (Tma12) with high sequence similarity to LPMOs from the AA10 family was identified in the fern Tectaria macrodonta [22]. Tma12 was identified through purification of extracts from fronds coupled with a reverse mass spectrometry approach. Interestingly, heterologous expression of Tma12 in cotton conferred an increased resistance to whitefly infection. This observation suggests a similar insecticidal effect of the T. macrodonta LPMO as the putative LPMO from the insect poxvirus [23]. The crystal structure was solved and showed structural similarity to cellulolytic LPMOs [19]. This indicates that LPMOs can be found in at least one branch of the plant kingdom.
LPMOs contain a Cu-histidine brace motif
The main feature of the LPMO structure is the metal-binding site, composed of two histidine residues exposing the bound Cu atom to the environment, situated in the middle of one of the flat surfaces of the protein later shown to bind substrate (Figure 1). This is the Cu-histidine brace motif. The two nitrogen atoms from the histidine side-chains and a third from the N-terminus are all interacting with the Cu-atom at a distance of close to 2 Å [9,24].
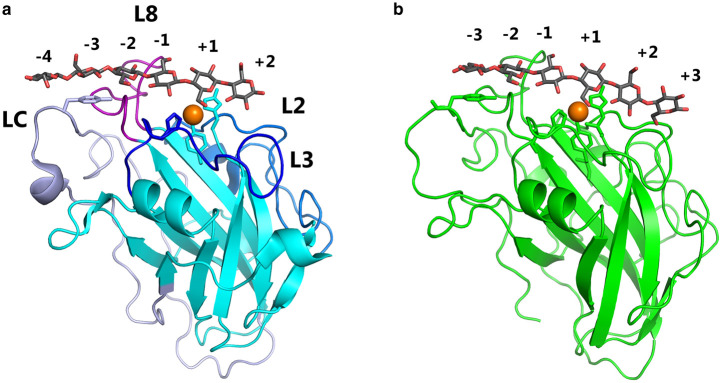
Figure 1. Structures of two LPMOs in complex with their substrate.
Interactions of LsAA9A (a) and CvAA9A (b) in crystal complexes with cellohexaose (PDB codes 5ACI and 6YDE [29,33]). Substrate-binding subsites are indicated by numbers. The active site copper is shown as an orange sphere, with interacting residues as sticks. The Tyr from the LC loop interacting with the substrate at the −3 subsite is also shown. Hydrogen bonding residues from the L2, L3 and L8 loops are not shown for clarity. Loops are indicated on the LsAA9A structure. Protein structures were visualised using PyMOL 2.4.
One of the first characterised LPMOs was found in Thermoascus aurantiacus (TaAA9A). The affinity for Cu(II) was found by isothemal titration calorimetry (ITC) to be in the low nanomolar range, this was confirmed using electron paramagnetic resonance (EPR), which suggested a dissociation constant (Kd) in the pM range [9]. Similar direct determination of affinity has been carried out for an AA10 member from Serratia marcescens (SmAA10A). Interestingly, SmAA10A was found to bind Zn(II) in addition to Cu(II). The experimentally determined Kd of 55 nM for Cu(II)-SmAA10A was used to estimate Kd of 1.2 nM for Cu(I)-SmAA9A [25]. This study clearly showed that LPMOs have a higher affinity for the reduced metal Cu(I). The fact that certain LPMO families can bind Zn(II) was utilised to accurately determine the affinity of Aspergillus oryzae AA11 (AoAA11) towards copper. Displacement ITC using (Zn(II)) as the weak ligand to be displaced by the stronger ligand (Cu(II)), found a Kd of 0.7 nM for AoAA11 [26] and of 43 nM for Bacillus amyloliquefaciens AA10 [27]. Taken together, these studies demonstrate a very high affinity of LPMO families towards copper. Interestingly, it has been demonstrated that LPMOs have an increased affinity for polysaccharide substrate when reduced [28]. This could mean that the reduction in the metal not only primes the enzyme for catalysis, but also directs it towards its substrate by increased affinity.
Structural features of LPMOs
Determination of the structures of LPMOs has contributed considerably to our understanding of LPMOs (detailed structural reviews can be found in [29–31]). To date, structures are available for over 40 different LPMOs, belonging primarily to the AA9 and AA10 families, and one representative each of the AA11, AA13, AA14 and AA15 families. Common to all LPMOs is a β-sandwich core structure consisting of two β-sheets comprising seven or eight β-strands in total. Structural diversity is generated by the helices and loops that connect the core β-strands, giving rise to variations in the dimensions and topologies of the substrate-binding surface. Fungal LPMOs are often methylated at the N-terminal histidine. The role of this methylation was recently investigated through parallel assays of TaAA9A expressed in Pichia pastoris (unmethylated) and Aspergillus oryzae (methylated), and it was suggested that methylation is involved in protection against auto-oxidative inactivation of the LPMO [32]. The reason why methylation is only observed in LPMOs expressed in filamentous fungi is not known.
The interaction between LPMOs and their polysaccharide substrate
When the first structures of fungal LPMOs in the AA9 family were determined, it became apparent that they shared a striking structural similarity to certain carbohydrate-binding modules (CBMs) [17,34,35]. This confirmed a connection between the AA9 and AA10 families (formerly GH61 and CBM33). Some features were lacking compared with glycoside hydrolases, most notably a clear active site cleft or groove. These absences pointed towards a different reaction mechanism than the one identified in glycoside hydrolases. However, some similarities were found as an AA9 LPMO from the ascomycete Thielavia terrestris (now Thermothielavioides terrestris) showed an arrangement of aromatic residues that was strongly reminiscent of a carbohydrate binding module with specificity for crystalline polysaccharides [36], suggesting that these proteins acted by binding to crystalline cellulose.
It has been difficult to obtain detailed structural information on the interaction of LPMOs with their polysaccharide substrates [13,37]. However, X-ray crystallographic, nuclear magnetic resonance (NMR), EPR studies, and computational studies have given us some insights into the process. Interaction studies by crystallography are only possible on LPMOs able to bind relatively small fragments (oligosaccharides), and it has so far only been successful with two AA9 enzymes that are able to cleave oligosaccharides by C4 oxidation [29,33,38]. In addition to the His-brace region, four loops contribute residues interacting with the substrate, mostly through hydrogen bonds, but also with a highly conserved aromatic residue (Tyr203 in LsAA9A, Figure 1a). These studies have provided a very detailed picture of the interactions of the active sites of these LPMOs with their oligosaccharide substrates. Unfortunately, these insights are not directly transferable to LPMOs acting on crystalline polysaccharides.
Another experimental technique that can provide structural information on the interaction of LPMOs with substrates is NMR spectroscopy, which can be used to identify protein residues interacting with the polysaccharide through changes in chemical shifts, as carried out for both AA9 and AA10 LPMOs [25,39]. However, NMR is only suitable for diamagnetic species, and the paramagnetic Cu(II) will distort signals from all nearby nuclei. Thus, this technique is useful when the LPMO is in e.g. Cu(I), Zn(II) or apo-form. While NMR spectroscopy cannot provide the same level of detail as X-ray crystallography, it has the advantage that interactions with both large polysaccharide substrates and small oligosaccharides can be probed.
EPR spectroscopy takes advantage of the paramagnetic properties of Cu(II), and has been used extensively in LPMO research to probe the active-site copper. EPR spectroscopy has recently been utilised to provide experimental information in the modelling of interactions between LPMO and oriented celery cellulose fibres [40], confirming previous suggestions that LsAA9A attacks the edge of cellulose fibres, rather than the flatter surfaces. Surprisingly, it was concluded that TaAA9A also attacks the same edge, which is somewhat counterintuitive given the rather different shapes of the binding surfaces of these two LPMOs.
Computational studies (docking and molecular dynamics) have been carried out to investigate the interactions of LPMOs with crystalline polysaccharides; for example, an extensive study on chitinolytic AA10 [41], and the recent study on a cellulose-specific LPMO [42], in both cases showing interactions with the flat faces of crystalline polysaccharides.
LPMOs use the oxidative power of oxygen
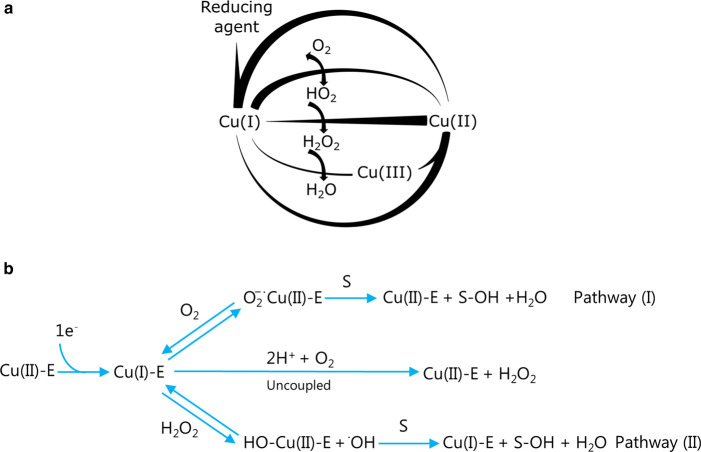
LPMOs have been classified as oxidases by the International Union of Biochemistry and Molecular Biology, and their catalytic reaction with O2 and polysaccharides gives the numbers EC 1.14.99.53 (chitin), EC 1.14.99.54 (C1 dehydrogenation on cellulose), EC 1.14.99.56 (C4 dehydrogenation on cellulose), and EC 1.14.99.55 (starch). However, the current understanding of the catalytic competencies of LPMOs is far more complex and is briefly discussed here. Figure 2a illustrates Cu-catalysed reactions coupled to the reduction in O2 to water at circumneutral pH [43,44]. The reduction in O2 to superoxide is thermodynamically disfavoured, and only trace amounts of superoxide are produced. Superoxide is reduced by a fast and efficient reaction with Cu(I) to form H2O2, and it is this fast reduction in superoxide that drives the reduction in O2 by virtue of Le Chatelier's principle. The surface-exposed Cu-histidine brace of LPMOs can take part in similar chemical reactions. The simplified LPMO reaction scheme shown in Figure 2b illustrates this point well. In pathway I, the resting-state LPMO (Cu(II)-E) is first reduced by one electron to Cu(I)-E, which is re-oxidised by O2. The reaction has been experimentally demonstrated, and the re-oxidation rate found to differ 10 fold between two LPMOs [45,46]. An uncoupled reaction that can be detected by the production of H2O2 takes place in the absence of a polysaccharide substrate [45,47–49]. The mechanism behind the uncoupled reaction has been investigated using quantum mechanics/molecular mechanics simulations [50,51]. In the presence of a polysaccharide substrate the monooxygenase reaction takes place, and oxidised saccharide products and water are produced, while returning the enzyme to the resting state Cu(II)-E. This was first demonstrated using 18O2 for the chitin-active SmAA10A [10], and confirmed for a cellulose-active LPMO from Neurospora crassa (NCU08760) [52]. In pathway II, H2O2 reacts as the co-substrate with Cu(I)-E in a reaction that produces oxidised saccharide products, water and the reduced enzyme. The H2O2 may be from an external source or it may be produced by the LPMOs in the uncoupled reaction. Pathway II was first suggested in 2017 [49] and examined with a cellulolytic AA9 enzyme from Hypocrea jecorina [46]. The re-oxidation rate of Cu(I)-LPMO with H2O2 was found to be 1,000 times faster than with O2 [46,53]. The reaction path with H2O2 is likely branched, but homolytic cleavage of H2O2 has been identified as the predominant path based on single-turnover experiments [46].
Figure 2. Simplified reaction schemes for free copper and Cu-LPMO.
(a) Free copper reactions based on [44] (b) Cu-LPMO reactions based on [31,48]. Pathway I: O2 binds to the Cu(I)-active site of the Enzyme (E). In the presence of a polysaccharide substrate (S), O2 reduction leads to substrate hydroxylation. In the absence of a polysaccharide, O2 reduction produces H2O2. Pathway II: H2O2 generated in situ can react with the Cu(I)-active site. When the polysaccharide is bound, substrate oxidation occurs. Small-molecule reductants or enzymes, such as cellobiose dehydrogenase, can provide the necessary reducing equivalents.
As described above, LPMOs bind poly- or oligosaccharides in a fixed position with no distortion of the substrate across the Cu-histidine brace [29]. It has been suggested that the binding of a polysaccharide and a co-substrate may be synergistic [29], random-sequential [48] or ordered-sequential [46]. Cleavage of the glycosidic bonds is believed to start with the abstraction of a hydrogen atom from carbon number 1 or 4 of the sugars (C1 or C4) on either side of the bond. This is a chemically difficult reaction because of the C-H bond strength of nearly 100 kcal/mol [54]. The regiospecificity (i.e. the propensity of the products to be oxidised at C1 or C4) is not strictly associated with AA families, and can be substrate-dependent [38,54,55]or reductant-dependent [56]. Although several mechanisms have been proposed, and some examined computationally [57], the mechanistic details of the reaction(s) leading to the cleavage of glycosidic bonds are still unknown.
LPMOs in biotechnological applications
While pure cellulose is not degraded by LPMOs in classic cellulase assays, it is clear that lignocellulose is [9]. The liquid fraction from pretreated lignocellulose, such as straw, contains different low-molecular-weight compounds that instigate activity. Many reducing agents, such as gallic acid [9], ascorbate, cysteine [18] and others [12,58] will have the same effect, and have been used in laboratory experiments. However, the relationship between reducing agents (including oxidoreductases and metals [9,14,52,59,60]), LPMOs and oxygen during the saccharification of lignocellulose is complicated [61,62]. Our own studies have shown that the industrial decomposition of lignocellulose takes place at low levels of dissolved O2, due primarily to abiotic O2-consuming reactions [61,62]. Importantly, we found that even when the lignocellulosic material was flushed with only 2% O2 (compared with air), reactive oxygen species had to be removed by catalase in order to stabilise the enzyme cocktail. This finding implies that H2O2 is generated during incubation of the material. As discussed above, H2O2 has been shown to function as a co-substrate in the place of O2 in a rapid catalytic cycle [53]. It was recently shown that the liquid fraction of pretreated wheat straw could provide both electrons and H2O2 for LPMO catalysis under oxic conditions [63]. Evidence suggests that while H2O2 leads to faster initial rates of polysaccharide oxidation than O2 [46], it also reduces the half-life of the enzyme cocktail in industrially relevant assays [61]. Furthermore, oxic conditions also resulted in acidification and decarboxylation of the lignocellulosic material by chemical reactions that are affected by oxidoreductases in the mixture [62]. Abiotic and enzymatic oxidative processes are thus intertwined during the decomposition of plant materials, and transition metals, Cu in particular, are involved in both.
LPMOs and the industrial use of these enzymes have been described in the patent literature. A large proportion of patent applications describe technical advances of relevance to biotechnological processes involving plant material. The use of LPMOs for the decomposition of cellulose [17] and starch [64] are good examples thereof. In another example, the inventors claimed the use of an AA9 LPMO that has activity on xylan [65]. Xylans are major components of hemicellulose involved in the integrity of the plant cell wall. The removal of xylans increases the productivity of commercial enzymes, likely by increasing substrate accessibility [66]. LPMOs may also improve the fermentation of starch-derived sugars to desirable products by reducing the formation of lactic or acetic acidic acid during incubation [67]. Several patent applications that mainly disclose DNA sequences of several amino acid variants of LPMOs with desirable qualities have been published, a recent example being that by Lin et al. [68]. These are a few examples from the patent landscape that document the broad commercial interest and potentially significant economic gain of using LPMOs as biotechnological process tools.
LPMO-like proteins and other proteins that share structural features with LPMOs
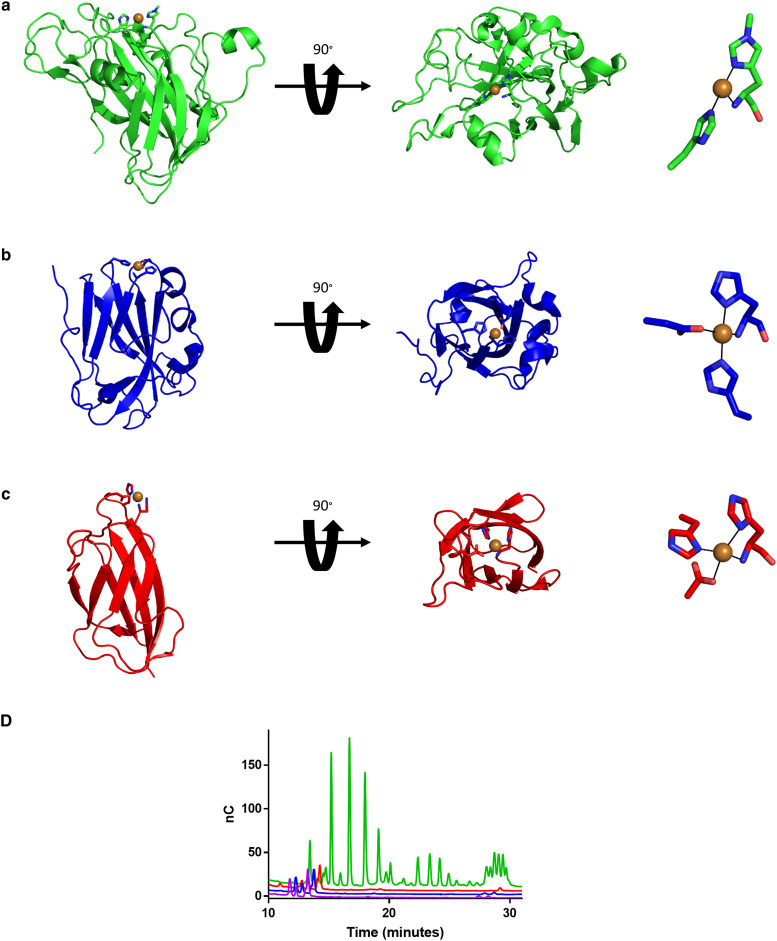
As discussed above, the field of LPMO research has historically had a strong focus on enzymes with potential to be applied in biomass reactors. The structure of a model cellulose active LPMO TaAA9A is shown in Figure 3a. More recently, the field has turned towards exploring the roles and diversity of these enzymes in nature. However, the identification of LPMOs has proven to be more difficult than first expected. One attempt to identify novel fungal LPMOs instead revealed a family of LPMO-like proteins (termed X325). The X-ray structure of a member of the X325 family from the ectomycorrhizal fungus Laccaria bicolor has been solved (Figure 3b) [37]. Although the structure displays a His brace Cu-binding site on the protein, no polysaccharide-degrading activity has been detected for several members of X325 [37,69]. One member of the X325 family has been identified in the human pathogen Cryptococcus neoformans. The expression of this protein, called Bim1, was seen to dramatically increase under Cu-limiting conditions. Bim1 participates in Cu uptake in concert with the Cu(I) importer Ctr1, and is a critical factor for Cu acquisition in fungal meningitis [69]. Intriguingly, a similar Cu-His-brace motif has also been found in the small bacterial periplasmic protein CopC (Figure 3c), which also functions in copper homeostasis and the delivery of copper to specific transporters. In a recent study, we compared the biochemical properties of CopC from Pseudomonas fluorescens with Bim1 and the well-described cellulose-specific TaAA9A [44]. Only TaAA9 was found to cleave cellulose (Figure 3d) as was expected because it contains an appropriate polysaccharide binding site while the others do not. However, the CopC protein is inert to the relevant reductant ascorbate [44] and thus unable to initiate the cycle between redox states that is associated with LPMO activity. This confirms that CopC is not an LPMO despite the presence of a Cu histidine brace. The importance of some first and second sphere residues in LPMOs for activity has been investigated experimentally [17,70] and computationally [57,71]. These important residues could explain the differences in copper reactivity. For a more in depth review of the hydrogen bonding network and implications of the second coordination sphere see [72].
Figure 3. Three protein folds all containing the Cu-histidine brace.
A(a) Representative cellulose-active LPMO, TaAA9A (green) (PDB: 2YET) [9]. (b) The LPMO-like protein LaX325 (blue) (PDB: 6IBJ) from Laetisaria arvalis which, based on sequence and predicted fold, belongs to the same family of LPMO-like proteins as Bim1 [37]. (c) The Cu chaperone, CopC, from Pseudomonas fluorescence (red) (PDB: 6NFQ) [75]. (d) Results of HPAEC-PAD analysis of the three proteins when incubated with phosphoric acid swollen cellulose and ascorbate for 24 h [44]. Only the LPMO trace (green) shows polysaccharide-degrading activity. The purple trace is that of aqueous copper. Protein structures were visualised using PyMOL 2.4.
Another Cu-histidine brace found in nature is the B-site in particulate methane monooxygenases. These enzymes catalyse the oxidation of methane to methanol. Until recently, this site was identified as the active site, in part due to its similarities to the LPMO histidine brace. However, another mononuclear copper site with different coordination geometry more recently has been suggested to be the active site [73,74]. It is thus clear that biochemical demonstration of substrate cleavage activity is important when claiming redox activity, since both the LPMO fold and Cu-histidine brace motif can adapt to different and diverse functions.
Perspectives
Within the field of industrial biotechnology, LPMOs are of the utmost importance for efficient decomposition and processing of plant materials.
LPMOs and LPMO-like proteins are widely found in nature. The Cu-active site of LPMOs undergoes redox-cycling to catalyse cleavage of polysaccharides using molecular mechanisms that rely on redox partners and use oxygen species as co-substrate.
Both enzymatic and abiotic oxidative processes must be carefully controlled in biorefineries. Further studies are required to shed further light on how LPMOs acts in vivo.
Acknowledgements
The authors thank Professor Poul Erik Jensen and Professor Meike Burow for their very significant contributions to the scientific environment.
Abbreviations
- CAZy
Carbohydrate-Active EnZymes database
- EPR
electron paramagnetic resonance
- ITC
isothemal titration calorimetry
- LPMOs
Lytic polysaccharide monooxygenases
- NMR
nuclear magnetic resonance
- PDB
Protein Data Bank
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This study was supported by the Novo Nordisk Foundation, Grant No. NNF17SA0027704 to K.S.J.
Author Contribution
All authors were involved with writing the paper.
References
- 1.Johansen, K.S. (2016) Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem. Soc. Trans. 44, 143–149 10.1042/BST20150204 [DOI] [PubMed] [Google Scholar]
- 2.Maersk join forces with industry peers and customers to develop LEO [press release]. https://www.maersk.com/news/articles/2019/10/29/maersk-join-forces-with-industry-peers-and-customers-to-develop-leo: Maersk, A.P. Moller -, 29. October 2019
- 3.Reese, E.T. (1956) Enzymatic hydrolysis of cellulose. Appl. Microbiol. 4, 39–45 10.1128/AM.4.1.39-45.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boisset, C., Pétrequin, C., Chanzy, H., Henrissat, B. and Schülein, M. (2001) Optimized mixtures of recombinant Humicola insolens cellulases for the biodegradation of crystalline cellulose. Biotechnol. Bioeng. 72, 339–345 [DOI] [PubMed] [Google Scholar]
- 5.Tokin, R., Ipsen, J.Ø, Westh, P. and Johansen, K.S. (2020) The synergy between LPMOs and cellulases in enzymatic saccharification of cellulose is both enzyme- and substrate-dependent. Biotechnol. Lett. 42, 1975–1984 10.1007/s10529-020-02922-0 [DOI] [PubMed] [Google Scholar]
- 6.Vaheri, M.P. (1982) Acidic degradation products of cellulose during enzymatic hydrolysis by Trichoderma reesei. J. Appl. Biochem. 4, 153–160 [Google Scholar]
- 7.Vaheri, M.P. (1982) Oxidation as a part of degradation of crystalline cellulose by Trichoderma reesei. J. Appl. Biochem. 4, 356–363 [Google Scholar]
- 8.Eriksson, K.-E., Pettersson, B. and Westermark, U. (1974) Oxidation: an important enzyme reaction in fungal degradation of cellulose. FEBS Lett. 49, 282–285 10.1016/0014-5793(74)80531-4 [DOI] [PubMed] [Google Scholar]
- 9.Quinlan, R.J., Sweeney, M.D., Lo Leggio, L., Otten, H., Poulsen, J.C., Johansen, K.S.et al. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl Acad. Sci. U.S.A. 108, 15079–15084 10.1073/pnas.1105776108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaaje-Kolstad, G., Westereng, B., Horn, S.J., Liu, Z., Zhai, H., Sørlie, M.et al. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- 11.Levasseur, A., Drula, E., Lombard, V., Coutinho, P.M. and Henrissat, B. (2013) Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6, 41 10.1186/1754-6834-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frommhagen, M., Sforza, S., Westphal, A.H., Visser, J., Hinz, S.W.A., Koetsier, M.J.et al. (2015) Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol. Biofuels 8, 101 10.1186/s13068-015-0284-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borisova, A.S., Isaksen, T., Dimarogona, M., Kognole, A.A., Mathiesen, G., Várnai, A.et al. (2015) Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J. Biol. Chem. 290, 22955–22969 10.1074/jbc.M115.660183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Leggio, L., Simmons, T.J., Poulsen, J.-C.N., Frandsen, K.E.H., Hemsworth, G.R., Stringer, M.A.et al. (2015) Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat. Commun. 6, 5961 10.1038/ncomms6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monclaro, A.V., Petrović, D.M., Alves, G.S.C., Costa, M.M.C., Midorikawa, G.E.O., Miller, R.N.G.et al. (2020) Characterization of two family AA9 LPMOs from Aspergillus tamarii with distinct activities on xyloglucan reveals structural differences linked to cleavage specificity. PLoS ONE 15, e0235642 10.1371/journal.pone.0235642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limsakul, P., Phitsuwan, P., Waeonukul, R., Pason, P., Tachaapaikoon, C., Poomputsa, K.et al. (2020) A novel AA10 from Paenibacillus curdlanolyticus and its synergistic action on crystalline and complex polysaccharides. Appl. Microbiol. Biotechnol. 104, 7533–7550 10.1007/s00253-020-10758-x [DOI] [PubMed] [Google Scholar]
- 17.Harris, P.V., Welner, D., McFarland, K.C., Re, E., Navarro Poulsen, J.C., Brown, K.et al. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49, 3305–3316 10.1021/bi100009p [DOI] [PubMed] [Google Scholar]
- 18.Couturier, M., Ladevèze, S., Sulzenbacher, G., Ciano, L., Fanuel, M., Moreau, C.et al. (2018) Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat. Chem. Biol. 14, 306 10.1038/nchembio.2558 [DOI] [PubMed] [Google Scholar]
- 19.Yadav, S.K., Archana, Singh, R., Singh, P.K. and Vasudev, P.G. (2019) Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase. Planta 249, 1987–1996 10.1007/s00425-019-03135-0 [DOI] [PubMed] [Google Scholar]
- 20.Sabbadin, F., Hemsworth, G.R., Ciano, L., Henrissat, B., Dupree, P., Tryfona, T.et al. (2018) An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion. Nat. Commun. 9, 756 10.1038/s41467-018-03142-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filiatrault-Chastel, C., Navarro, D., Haon, M., Grisel, S., Herpoël-Gimbert, I., Chevret, D.et al. (2019) AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol. Biofuels 12, 55 10.1186/s13068-019-1394-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla, A.K., Upadhyay, S.K., Mishra, M., Saurabh, S., Singh, R., Singh, H.et al. (2016) Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 34, 1046–1051 10.1038/nbt.3665 [DOI] [PubMed] [Google Scholar]
- 23.Chiu, E., Hijnen, M., Bunker, R.D., Boudes, M., Rajendran, C., Aizel, K.et al. (2015) Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization. Proc. Natl Acad. Sci. U.S.A. 112, 3973–3978 10.1073/pnas.1418798112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciano, L., Davies, G.J., Tolman, W.B. and Walton, P.H. (2018) Bracing copper for the catalytic oxidation of C–H bonds. Nat. Catal. 1, 571–577 10.1038/s41929-018-0110-9 [DOI] [Google Scholar]
- 25.Aachmann, F.L., Sørlie, M., Skjåk-Bræk, G., Eijsink, V.G.H. and Vaaje-Kolstad, G. (2012) NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proc. Natl Acad. Sci. U.S.A. 109, 18779–18784 10.1073/pnas.1208822109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemsworth, G.R., Henrissat, B., Davies, G.J. and Walton, P.H. (2014) Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat. Chem. Biol. 10, 122–126 10.1038/nchembio.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory, R.C., Hemsworth, G.R., Turkenburg, J.P., Hart, S.J., Walton, P.H. and Activity, D.G. (2016) Stability and 3-D structure of the Cu(II) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens. Dalton Trans. 45, 16904–16912 10.1039/C6DT02793H [DOI] [PubMed] [Google Scholar]
- 28.Kracher, D., Andlar, M., Furtmüller, P.G. and Ludwig, R. (2018) Active-site copper reduction promotes substrate binding of fungal lytic polysaccharide monooxygenase and reduces stability. J. Biol. Chem. 293, 1676–1687 10.1074/jbc.RA117.000109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frandsen, K.E.H. and Lo Leggio, L. (2016) Lytic polysaccharide monooxygenases: a crystallographer's view on a new class of biomass-degrading enzymes. IUCrJ 3, 448–467 10.1107/S2052252516014147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaaje-Kolstad, G., Forsberg, Z., Loose, J.S., Bissaro, B. and Eijsink, V.G. (2017) Structural diversity of lytic polysaccharide monooxygenases. Curr. Opin. Struct. Biol. 44, 67–76 10.1016/j.sbi.2016.12.012 [DOI] [PubMed] [Google Scholar]
- 31.Tandrup, T., Frandsen, K.E.H., Johansen, K.S., Berrin, J.G. and Lo Leggio, L. (2018) Recent insights into lytic polysaccharide monooxygenases (LPMOs). Biochem. Soc. Trans. 46, 1431–1447 10.1042/BST20170549 [DOI] [PubMed] [Google Scholar]
- 32.Petrović, D.M., Bissaro, B., Chylenski, P., Skaugen, M., Sørlie, M., Jensen, M.S.et al. (2018) Methylation of the N-terminal histidine protects a lytic polysaccharide monooxygenase from auto-oxidative inactivation. Protein Sci. 27, 1636–1650 10.1002/pro.3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tandrup, T., Tryfona, T., Frandsen, K.E.H., Johansen, K.S., Dupree, P. and Lo Leggio, L. (2020) Oligosaccharide binding and thermostability of two related AA9 lytic polysaccharide monooxygenases. Biochemistry 59, 3347–3358 10.1021/acs.biochem.0c00312 [DOI] [PubMed] [Google Scholar]
- 34.Karkehabadi, S., Hansson, H., Kim, S., Piens, K., Mitchinson, C. and Sandgren, M. (2008) The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 Å resolution. J. Mol. Biol. 383, 144–154 10.1016/j.jmb.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 35.Welner, D. (2006) Four Proteins in Glycobiology: Structure and Function, University of Copenhagen [Google Scholar]
- 36.Boraston, A.B., Bolam, D.N., Gilbert, H.J. and Davies, G.J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 10.1042/BJ20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labourel, A., Frandsen, K., Zhang, F., Brouilly, N., Grisel, S., Haon, M.et al. (2020) A fungal family of lytic polysaccharide monooxygenase-like copper proteins. Nat. Chem. Biol. 16, 345–350 10.1038/s41589-019-0438-8 [DOI] [PubMed] [Google Scholar]
- 38.Simmons, T.J., Frandsen, K.E.H., Ciano, L., Tryfona, T., Lenfant, N., Poulsen, J.C.et al. (2017) Structural and electronic determinants of lytic polysaccharide monooxygenase reactivity on polysaccharide substrates. Nat. Commun. 8, 1064 10.1038/s41467-017-01247-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courtade, G., Wimmer, R., Røhr, Å.K, Preims, M., Felice, A.K.G., Dimarogona, M.et al. (2016) Interactions of a fungal lytic polysaccharide monooxygenase with β-glucan substrates and cellobiose dehydrogenase. Proc. Natl Acad. Sci. U.S.A. 113, 5922–5927 10.1073/pnas.1602566113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciano, L., Paradisi, A., Hemsworth, G.R., Tovborg, M., Davies, G.J. and Walton, P.H. (2020) Insights from semi-oriented EPR spectroscopy studies into the interaction of lytic polysaccharide monooxygenases with cellulose. Dalton Trans. 49, 3413–3422 10.1039/C9DT04065J [DOI] [PubMed] [Google Scholar]
- 41.Bissaro, B., Isaksen, I., Vaaje-Kolstad, G., Eijsink, V.G.H. and Røhr, Å.K. (2018) How a lytic polysaccharide monooxygenase binds crystalline chitin. Biochemistry 57, 1893–1906 10.1021/acs.biochem.8b00138 [DOI] [PubMed] [Google Scholar]
- 42.Zhou, H., Zhang, Y., Li, T., Tan, H., Li, G. and Yin, H. (2020) Distinct interaction of lytic polysaccharide monooxygenase with cellulose revealed by computational and biochemical studies. J. Phys. Chem. Lett. 11, 3987–3992 10.1021/acs.jpclett.0c00918 [DOI] [PubMed] [Google Scholar]
- 43.Pham, A.N., Xing, G., Miller, C.J. and Waite, T.D. (2013) Fenton-like copper redox chemistry revisited: hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J. Catal. 301, 54–64 10.1016/j.jcat.2013.01.025 [DOI] [Google Scholar]
- 44.Brander, S., Horvath, I., Ipsen, J.Ø, Peciulyte, A., Olsson, L., Hernández-Rollán, C.et al. (2020) Biochemical evidence of both copper chelation and oxygenase activity at the histidine brace. Sci. Rep. 10, 16369 10.1038/s41598-020-73266-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kjaergaard, C.H., Qayyum, M.F., Wong, S.D., Xu, F., Hemsworth, G.R., Walton, D.J.et al. (2014) Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proc. Natl Acad. Sci. U.S.A. 111, 8797–8802 10.1073/pnas.1408115111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones, S.M., Transue, W.J., Meier, K.K., Kelemen, B. and Solomon, E.I. (2020) Kinetic analysis of amino acid radicals formed in H2O2-driven Cu(I) LPMO reoxidation implicates dominant homolytic reactivity. Proc. Natl Acad. Sci. U.S.A. 117, 11916–11922 10.1073/pnas.1922499117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kittl, R., Kracher, D., Burgstaller, D., Haltrich, D. and Ludwig, R. (2012) Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels 5, 79 10.1186/1754-6834-5-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hangasky, J.A., Iavarone, A.T. and Marletta, M.A. (2018) Reactivity of O2 versus H2O2 with polysaccharide monooxygenases. Proc. Natl Acad. Sci. U.S.A. 115, 4915–4920 10.1073/pnas.1801153115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bissaro, B., Røhr, Å.K, Müller, G., Chylenski, P., Skaugen, M., Forsberg, Z.et al. (2017) Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat. Chem. Biol. 13, 1123 10.1038/nchembio.2470 [DOI] [PubMed] [Google Scholar]
- 50.Caldararu, O., Oksanen, E., Ryde, U. and Hedegard, E.D. (2019) Mechanism of hydrogen peroxide formation by lytic polysaccharide monooxygenase. Chem. Sci. 10, 576–586 10.1039/C8SC03980A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, B., Walton, P.H. and Rovira, C. (2019) Molecular mechanisms of oxygen activation and hydrogen peroxide formation in lytic polysaccharide monooxygenases. ACS Catal. 9, 4958–4969 10.1021/acscatal.9b00778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips, C.M., Beeson, W.T., Cate, J.H. and Marletta, M.A. (2011) Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 10.1021/cb200351y [DOI] [PubMed] [Google Scholar]
- 53.Bissaro, B., Streit, B., Isaksen, I., Eijsink, V.G.H., Beckham, G.T., DuBois, J.L.et al. (2020) Molecular mechanism of the chitinolytic peroxygenase reaction. Proc. Natl Acad. Sci. U.S.A. 117, 1504–1513 10.1073/pnas.1904889117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hedegård, E.D. and Ryde, U. (2017) Targeting the reactive intermediate in polysaccharide monooxygenases. J. Biol. Inorg. Chem. 22, 1029–1037 10.1007/s00775-017-1480-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fanuel, M., Garajova, S., Ropartz, D., McGregor, N., Brumer, H., Rogniaux, H.et al. (2017) The podospora anserina lytic polysaccharide monooxygenase PaLPMO9H catalyzes oxidative cleavage of diverse plant cell wall matrix glycans. Biotechnol. Biofuels 10, 63 10.1186/s13068-017-0749-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cannella, D., Möllers, K.B., Frigaard, N.U., Jensen, P.E., Bjerrum, M.J., Johansen, K.S.et al. (2016) Light-driven oxidation of polysaccharides by photosynthetic pigments and a metalloenzyme. Nat. Commun. 7, 11134 10.1038/ncomms11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hedegård, E.D. and Ryde, U. (2018) Molecular mechanism of lytic polysaccharide monooxygenases. Chem. Sci. 9, 3866–3880 10.1039/C8SC00426A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, F., Sweeney, M., Quinlan, J. and Johansen, K.S. (2012) Novozymes (Novo-C) assignee. Aqueous composition useful for degrading or converting a cellulosic material, producing a fermentation product, and fermenting a cellulosic material comprises a polypeptide having cellulolytic enhancing activity and an organic compound patent WO2012021396-A1; US2013183723-A1; US8846351-B2. WO2012021396-A1 16 Feb 2012 C12P-007/10 201216
- 59.Beeson, W.T., Phillips, C.M., Cate, J.H.D. and Marletta, M.A. (2012) Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134, 890–892 10.1021/ja210657t [DOI] [PubMed] [Google Scholar]
- 60.Kracher, D., Scheiblbrandner, S., Felice, A.K.G., Breslmayr, E., Preims, M., Ludwicka, K.et al. (2016) Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352, 1098–1101 10.1126/science.aaf3165 [DOI] [PubMed] [Google Scholar]
- 61.Scott, B.R., Huang, H.Z., Frickman, J., Halvorsen, R. and Johansen, K.S. (2016) Catalase improves saccharification of lignocellulose by reducing lytic polysaccharide monooxygenase-associated enzyme inactivation. Biotechnol. Lett. 38, 425–434 10.1007/s10529-015-1989-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peciulyte, A., Samuelsson, L., Olsson, L., McFarland, K.C., Frickmann, J., Østergård, L.et al. (2018) Redox processes acidify and decarboxylate steam-pretreated lignocellulosic biomass and are modulated by LPMO and catalase. Biotechnol. Biofuels 11, 165 10.1186/s13068-018-1159-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kont, R., Pihlajaniemi, V., Borisova, A.S., Aro, N., Marjamaa, K., Loogen, J.et al. (2019) The liquid fraction from hydrothermal pretreatment of wheat straw provides lytic polysaccharide monooxygenases with both electrons and H2O2 co-substrate. Biotechnol. Biofuels 12, 235 10.1186/s13068-019-1578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soong, C., Saunders, J., Soong, C.L. and Song, Z. (2011) Novozymes North America Inc (Novo-C) Novozymes North America Inc (Novo-C) Novozymes North America Inc (Novo-C), assignee. Preparing fermented product used in consumable alcohol e.g. beer and wine, by converting starch-containing material to dextrin with alpha-amylase, saccharifying dextrin to sugar, and fermenting sugar using fermenting organism patent WO2011123505-A1
- 65.Koetsier, M.J., Visser, J., Hinz, S.W.A., Kabel, M.A., Frommhagen, M., Gruppen, H.et al. (2016) Genencor Int Bv (Dupo-C) Genencor Int Bv (Dupo-C) Genencor Int Bv (Dupo-C), assignee. Degrading and/or modifying xylan in xylan comprising substrate, or preparing baking composition used as e.g. building block for biochemical or bio fuel, by contacting xylan comprising substrate with lytic polysaccharide monooxygenase patent WO2016142536-A1
- 66.Jeoh, T., Ishizawa, C.I., Davis, M.F., Himmel, M.E., Adney, W.S. and Johnson, D.K. (2007) Cellulase digestibility of pretreated biomass is limited by cellulose accessibility. Biotechnol. Bioeng. 98, 112–122 10.1002/bit.21408 [DOI] [PubMed] [Google Scholar]
- 67.Gaspar, A.R., Medina, V.G.G., Li, X. and Leite Mulder, K.C. (2019) Shows A, inventors; NOVOZYMES AS (NOVO-C) GASPAR A R (GASP-Individual), assignee. Reducing and/or preventing increase in lactic acid levels in biofuel fermentation system comprises introducing lytic polysaccharide monooxygenase (LPMO) polypeptide or enzyme composition comprising LPMO polypeptide to system patent WO2019083831-A1
- 68.Lin, J., Bohan, D., Maranta, M., Beresford, L., Lamsa, M., Sweeney, M.et al. (2019) Inventors; NOVOYZMES AS (NOVO-Non-standard) NOVOZYMES INC (NOVO-C), assignee. New variant useful in composition for degrading or converting cellulosic material, or producing fermentation product chosen from alcohol, alkane, cycloalkane, alkene, amino acid, gas, isoprene, ketone, organic acid, and polyketide patent US2019153414-A1
- 69.Garcia-Santamarina, S., Probst, C., Festa, R., Ding, C., Smith, A., Conklin, S.et al. (2020) A lytic polysaccharide monooxygenase-like protein functions in fungal copper import and meningitis. Nat. Chem. Biol. 16, 337–344 10.1038/s41589-019-0437-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Span, E.A., Suess, D.L.M., Deller, M.C., Britt, R.D. and Marletta, M.A. (2017) The role of the secondary coordination sphere in a fungal polysaccharide monooxygenase. ACS Chem. Biol. 12, 1095–1103 10.1021/acschembio.7b00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Dell, W.B., Agarwal, P.K. and Meilleur, F. (2017) Oxygen activation at the active site of a fungal lytic polysaccharide monooxygenase. Angew. Chem. Int. Ed. Engl. 56, 767–770 10.1002/anie.201610502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vu, V.V. and Ngo, S.T. (2018) Copper active site in polysaccharide monooxygenases. Coord. Chem. Rev. 368, 134–157 10.1016/j.ccr.2018.04.005 [DOI] [Google Scholar]
- 73.Ross, M.O., MacMillan, F., Wang, J., Nisthal, A., Lawton, T.J., Olafson, B.D.et al. (2019) Particulate methane monooxygenase contains only mononuclear copper centers. Science 364, 566–570 10.1126/science.aav2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ro, S.Y., Schachner, L.F., Koo, C.W., Purohit, R., Remis, J.P., Kenney, G.E.et al. (2019) Native top-down mass spectrometry provides insights into the copper centers of membrane-bound methane monooxygenase. Nat. Commun. 10, 2675 10.1038/s41467-019-10590-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Udagedara, S.R., Wijekoon, C.J.K., Xiao, Z., Wedd, A.G. and Maher, M.J. (2019) The crystal structure of the copC protein from Pseudomonas fluorescens reveals amended classifications for the CopC protein family. J. Inorg. Biochem. 195, 194–200 10.1016/j.jinorgbio.2019.03.007 [DOI] [PubMed] [Google Scholar]