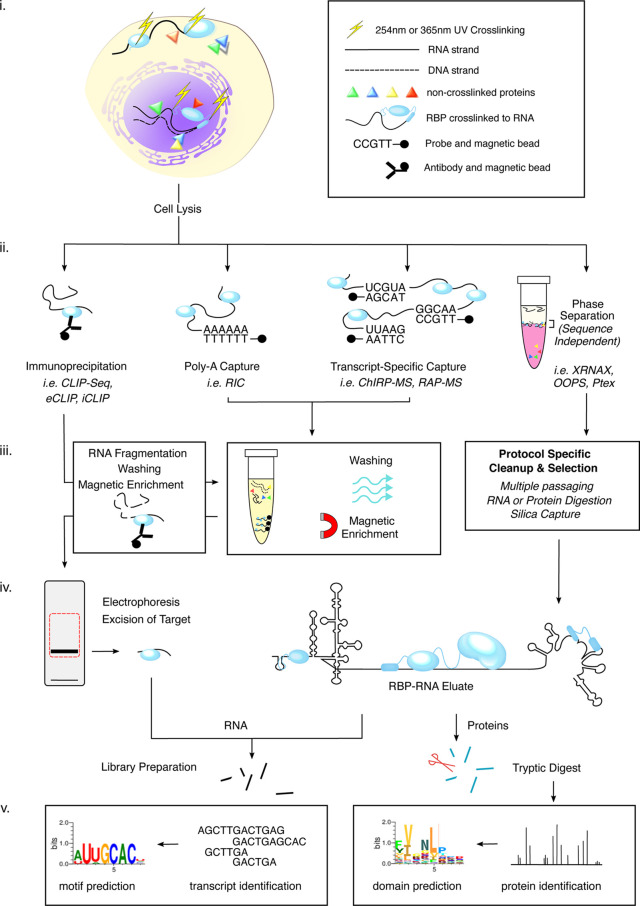
Figure 1. Common features of RBP–RNA capture methods.
(i) Intact cells are subjected to UV cross-linking of RBP to RNA either natively at 254 nm or, with assistance from 4SU, at 365 nm. (ii) The subsequent capture protocol will differ according to the study being conducted. From left to right. RBP-centric capture methods utilise immunoprecipitation to extract a population of target proteins in order to analyse their bound transcripts. RNA-centric methods such as poly-A capture or transcript specific capture use complementary probes to target an RNA compartment or RNA species, respectively. The former is frequently followed by global analysis of either RBP or RNA, where the latter is commonly used to investigate specific RBP interactors only. Phase separation methods capture interactions across the total transcriptome by first dissociating DNA, RNA and protein molecules. The cocktail's interphase is relied upon to enrich for protein–RNA complexes based on their compound physico-chemical character. (iii) Each capture method is subject to a clean-up step to improve sample purity. From left to right. Immunoprecipitants are subjected to RNA fragmentation and washing via magnetic separation. RNA-centric captures are also washed by magnetic separation although, because hybridisation anchors the transcript, these methods can tolerate higher stringencies than their immunoprecipitant counterparts. For phase separation methods clean-up steps can vary, even within a single protocol, depending on downstream application. Common aids are multiple passaging through the biphasic cocktail, RNA or protein digestion, and silica capture. In the case of Ptex, cocktail components can also vary. (iv) Immunoprecipitant methods often feature another round of separation by gel electrophoresis and products of a molecular mass close to the target recovered. The RBP–RNA complexes derived from RNA-centric and phase separation methods can be prepared for either RNA Seq or MS proteomics. (v) Sequencing of RNA yields can provide transcript identity or be used to identify RNA motifs based on the read densities flanking the cross-link sites. Similarly, MS proteomics can be used to identify and quantify the RBPs or a domain identification strategy used to find cross-link sites on the protein.