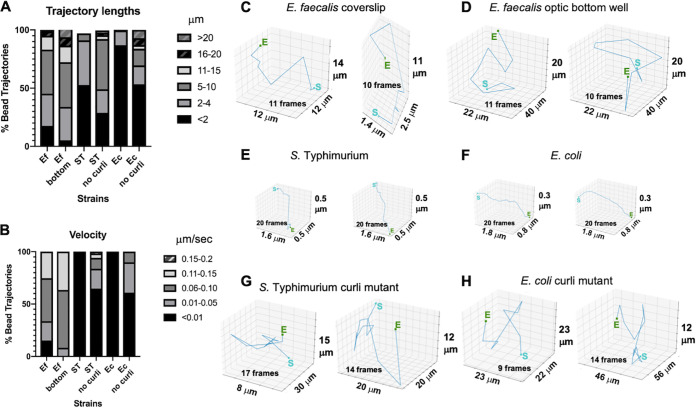
FIG 4.
The presence of curli shortens trajectory lengths and trajectory paths can be visualized for beads moving in biofilms lacking curli. The following biofilms were grown on coverslips: E. faecalis, S. Typhimurium, E. coli, and isogenic curli mutants, and E. faecalis was grown in an optical-bottom 96-well plate. (A) Trajectory lengths are given in micrometers and are presented as percentages of total bead trajectories. (B) Velocities are shown in micrometers per second and are presented as percentages of total bead trajectories. Two randomly chosen long trajectories with movement are visualized for each strain for E. faecalis (C), S. Typhimurium (E), E. coli (F), and isogenic curli mutants (G and H), or in an optical-bottom 96-well plate for E. faecalis (B). The sizes of the bounding boxes on the x, y, and z axes are shown. The boxes are not drawn to scale. The number of frames is noted in the bounding box. For beads with movement in the E. faecalis biofilms and Enterobacteriaceae curli mutant biofilms, the longest trajectories with movement were often less than 20 frames.