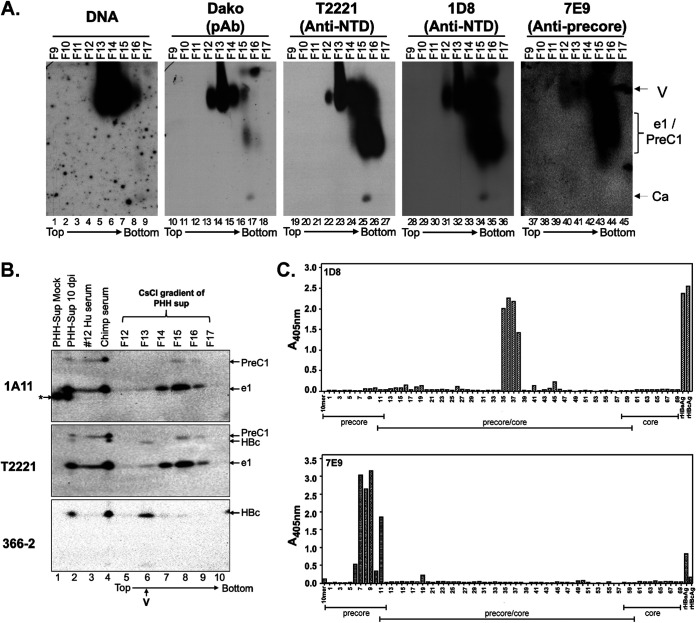
FIG 10.
Separation of HBeAg and PreC proteins from HBV virion particles secreted by HBV-infected PHH culture by CsCl density gradient fractionation. (A) Concentrated culture supernatant from gtD HBV-infected PHHs harvested 10 days postinfection (dpi) was fractionated by CsCl density gradient centrifugation. The virion particles and antigens in the fractions were analyzed by native agarose gel electrophoresis. HBV DNA and core protein were detected, as described in the legend to Fig. 6, using the indicated NTD- or CTD-specific MAbs. Ca, capsid, containing DNA or RNA or empty; e1/PreC1, secreted HBeAg and PreC protein from HBV genotype D-infected PHHs. (B) Concentrated culture supernatant from mock-infected or gtD HBV-infected PHHs harvested 10 days postinfection, as well as selected fractions from the CsCl gradient shown in panel A, were resolved by high-resolution SDS-PAGE, along with the serum from the gtD HBV-infected patient (no.12) and chimpanzee (no. 1616, week 22) (5; Hong et al., submitted). (C) Epitope mapping for MAbs 1D8 and 7E9 by ELISA using an overlapping peptide library. The linear epitope recognition of the anti-HBV precore/core MAbs 1D8 and 7E9 was interrogated against an overlapping, biotinylated peptide library, numbered 1 to 69, of the linear precore precursor protein (p25, residues −29 to 183 [Table 1]) epitopes, immobilized on a streptavidin-coated ELISA plate. Precore- and HBc-specific regions covered by the overlapping peptide library are indicated. Recombinant HBeAg and HBc were included as assay binding controls. The direction of centrifugation (top to bottom) is indicated in panels A and B.