FIG 13.
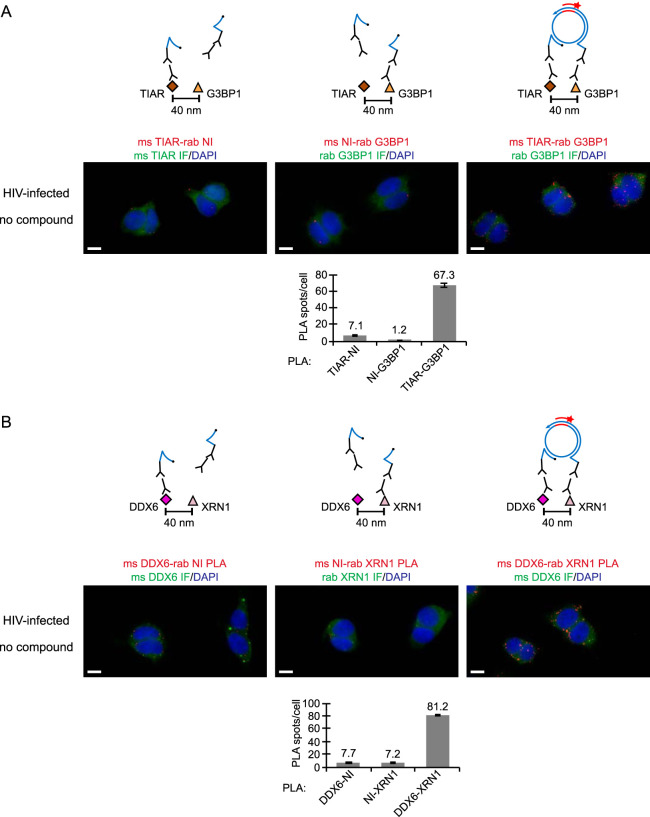
Nonimmune controls for the TIAR-G3BP1 and DDX6-XRN1 proximity ligation assays. (A) NI controls for the TIAR-G3BP1 PLA. Above each PLA image is the schematic corresponding to the PLA approach in that panel. Untreated 293T cells chronically infected with HIV-1 (293T-HIV) were analyzed by PLA. For the positive control (image and schematic on the right), PLA was performed by incubating with primary antibodies, mouse anti-TIAR and rabbit anti-G3BP1, followed by PLA secondary antibodies and other reagents as described in Fig. 6. In the two negative controls (images and schematics at left and center), one primary antibody was replaced with an NI control antibody from the same species, as indicated. Red spots indicating colocalization of the TIAR with G3BP should be absent when either primary antibody is replaced by an NI antibody. After PLA, TIAR or G3BP IF was performed to mark host protein-expressing cells with low-level green fluorescence. Images show a representative field for each of the three antibody pairs. Fields are shown as a merge of three color channels: the red channel shows TIAR-G3BP1 PLA, TIAR-NI PLA, or NI-G3BP1 PLA, as indicated by red labeling above images; the green channel shows TIAR IF or G3BP1 IF as indicated by green labeling above images; and the blue channel shows DAPI-stained nuclei. Scale bars, 10 μm. Graph below shows the average number of PLA spots per cell for each antibody pair. Five fields were analyzed for each group (containing a total of 45 to 62 cells per group), with error bars showing the SEM. (B) NI controls for the DDX6-XRN1 PLA. Above each PLA image is the schematic corresponding to the to the PLA approach in that panel. Untreated 293T cells chronically infected with HIV-1 (293T-HIV) were analyzed by PLA. For the positive control (image and schematic on the right), PLA was performed by incubating with primary antibodies, mouse anti-DDX6 and rabbit anti-XRN1, followed by PLA secondary antibodies and other reagents as described in Fig. 6. In the two negative controls (images and schematics at left and center), one primary antibody was replaced with an NI control antibody from the same species, as indicated. Red spots indicating colocalization of the DDX6 with XRN1 should be absent when either primary antibody is replaced by a NI antibody. Following PLA, DDX6 or XRN1 IF was performed to mark host protein-expressing cells with low-level green fluorescence. Images show a representative field for each of the three antibody pairs. Fields are shown as a merge of three color channels: the red channel shows DDX6-XRN1 PLA, DDX6-NI PLA, or NI-XRN1 PLA, as indicated by red labeling above images; the green channel shows DDX6 IF or XRN1 IF, as indicated by green labeling above images; and the blue channel shows DAPI-stained nuclei. Scale bars, 10 μm. Graph below shows the average number of PLA spots per cell for each antibody pair. Five fields were analyzed for each group (containing a total of 46 to 73 cells per group), with error bars showing the SEM.