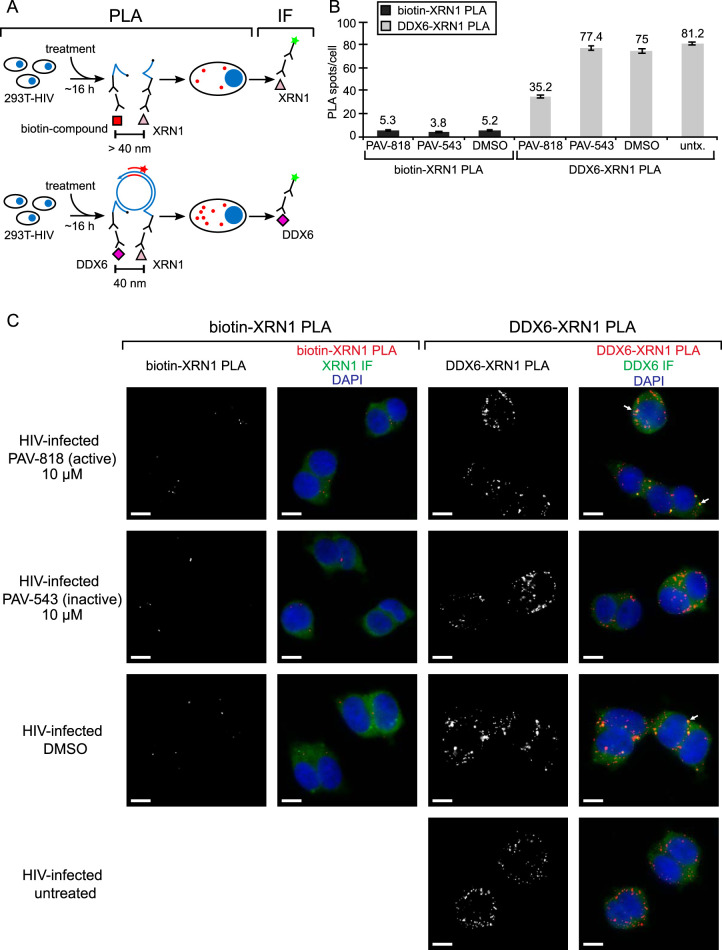
FIG 14.
The biotinylated antiretroviral PAV-206 analog does not colocalize in situ with XRN1, a host enzyme found in P-bodies. (A) Schematic of the PLA approaches for detecting colocalization of compound with XRN1 and colocalization of XRN1 with DDX6, both known to be present in P-bodies, are shown. 293T cells chronically infected with HIV-1 (293T-HIV) were treated with 10 μM PAV-818 (the biotinylated active compound), 10 μM PAV-543 (the biotinylated inactive compound), or DMSO for 16 h. The upper diagram depicts biotin-XRN1 PLA performed by incubation with primary antibodies (mouse anti-biotin and rabbit anti-XRN1), followed by PLA secondary antibodies and reagents as described in Fig. 6. Red spots indicate sites where biotinylated compound and XRN1 are colocalized in situ. After PLA, IF was performed (green star) to mark intracellular XRN1 with low-level green fluorescence. The lower diagram depicts DDX6-XRN1 PLA performed by incubation with primary antibodies (mouse anti-DDX6 and rabbit anti-XRN1), followed by PLA secondary antibodies as described as described in Fig. 6. Red spots indicate sites where the two P-body proteins DDX6 and XRN1 are colocalized in situ. After PLA, IF was performed (green star) to mark intracellular DDX6 with low-level green fluorescence. (B) The left side of the graph (dark gray bars) shows the average number of biotin-XRN1 PLA spots per cell for each condition, with representative images shown in panel C at the left. Five fields were analyzed for each group (containing a total of 44 to 54 cells per group), with error bars showing the SEM. The right side of the graph (light gray bars) shows the average number of DDX6-XRN1 PLA spots per cell for each condition, with representative images shown in panel C at the right. Five fields were analyzed for each group (containing a total of 57–73 cells per group), with error bars showing the SEM. (C) A representative field for each group quantified in the graph is shown. The two leftmost columns of images display biotin-XRN1 PLA spots alone in grayscale (first column) and a merge of three color channels (second column): biotin-XRN1 PLA (red), XRN1 IF (green), and DAPI-stained nuclei (blue). The two rightmost columns of images display DDX6-XRN1 PLA spots alone in grayscale (third column) and a merge of three color channels (fourth column): DDX6-XRN1 PLA (red), DDX6 IF (green), and DAPI-stained nuclei (blue). White arrows show sites where DDX6-XRN1 PLA signal colocalizes with large DDX6-IF-positive structures that are likely to be P-bodies. Scale bars, 10 μm.