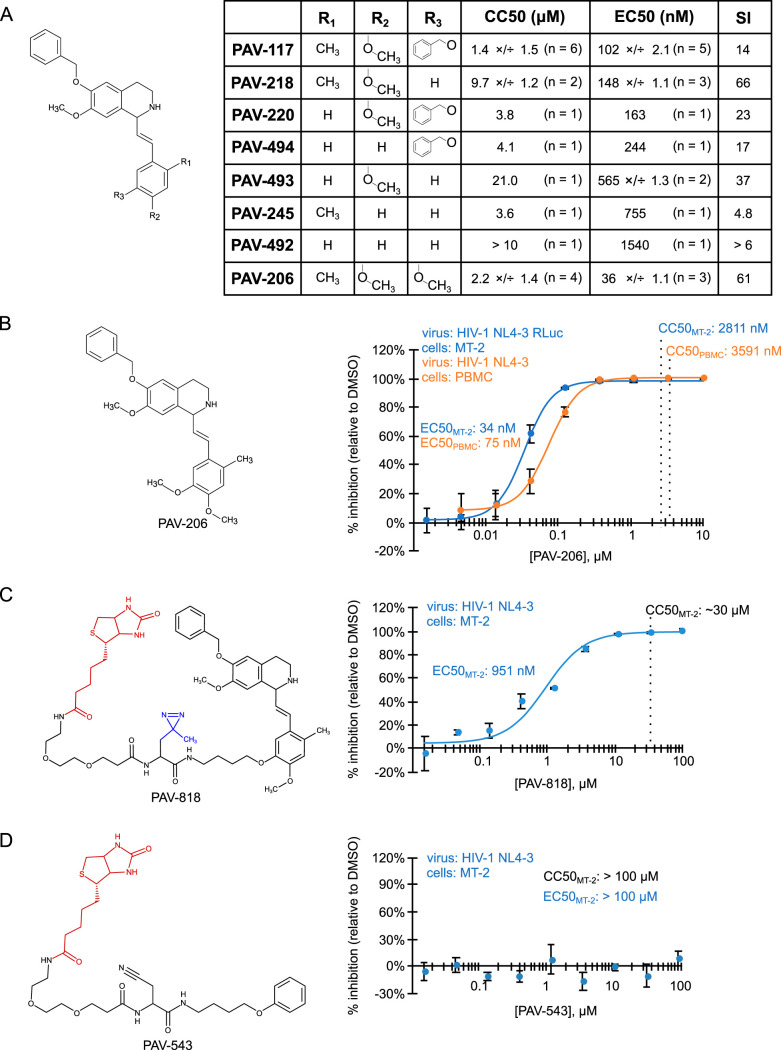
FIG 5.
Analysis of structure-activity relationships identified an analog that potently inhibits HIV-1 replication in PBMCs and a site for tags. (A) The general chemical structure of PAV-117 analogs is shown on the left, indicating the R1, R2, and R3 positions in the pendant benzene ring. The table shows results obtained for analogs in which the R1, R2, and R3 positions contain hydrogen, methyl, methoxy, or benzoyloxy groups as indicated, including the EC50 for inhibition of HIV-1 replication in MT-2 cells and the CC50 in MT-2 cells (assays described in Fig. 2C). Values are shown as the average of multiple independent repeats ×/÷ the GSD, with n = the number of independent repeats. Also shown is the selectivity index (SI), which is equivalent to CC50/EC50. (B) The structure of PAV-206 is shown on the left. The blue dose-response curve shows the inhibition of HIV-1 replication by PAV-206 in MT-2 T cells (using the assay described in Fig. 2C). The orange dose-response curve shows inhibition of HIV-1 replication by PAV-206 in PHA-activated PBMCs infected with unmodified HIV-1 NL4-3 at an MOI of 0.008. (C) On the left is the structure of PAV-818, the biotinylated analog of PAV-206, with the biotin moiety shown in red. Shown on the right is a dose-response curve for inhibition of HIV-1 replication by PAV-818 in MT-2 T cells (assay as in Fig. 2C). In the PAV-818 structure, a diazirine group is shown in blue. This group was added for future cross-linking studies but is not used in the present study. (D) Shown on the left is the structure of PAV-543, a biotinylated compound that does not have antiretroviral activity, with the biotin moiety in red. Shown on the right is a dose-response curve for inhibition of HIV-1 replication by PAV-543 in MT-2 T cells (assay as in Fig. 2C). For all graphs, the indicated EC50 values were determined from the dose-response curves. CC50 values were determined in uninfected cells in parallel and are marked by vertical dashed lines. Error bars in graphs show the SEM from three replicates.