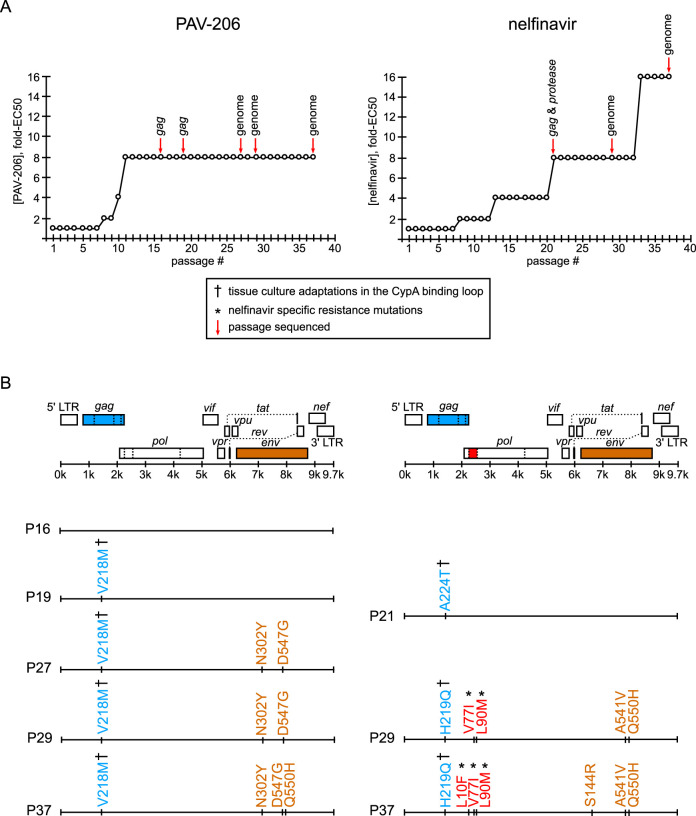
FIG 8.
Resistance mutations in gag or pol were not observed upon selection with PAV-206 in cell culture. (A) An NL4-3 virus stock was passaged weekly for 37 weeks in the presence of either PAV-206 or nelfinavir starting at a concentration of 1× EC50. The concentration of compound was increased 2-fold when compound-treated cultures reached maximum viral CPE in a time frame similar to a parallel DMSO control. The graph shows the compound concentration as the fold-EC50 versus week of selection (passage number) for PAV-206 (left) or nelfinavir (right). Red arrows indicate passages where virus was amplified, along with either sequencing of gag alone, gag and protease, or the full genome minus the LTRs (as indicated above the red arrows). (B) For each selection (PAV-206 versus nelfinavir), an HIV-1 genome map is shown, with positions of viral open reading frames (ORFs), 5′ LTR, and 3′ LTR, with a line below indicating their approximate nucleotide positions within the HIV-1 genome. In each case, ORFs in which dominant nonsynonymous mutations emerged during selection are color-coded as follows: gag, blue; pro, red; and env, brown. Beneath the HIV-1 genome maps, dominant nonsynonymous amino acid mutations (identified through sequencing of passages indicated by red arrows in panel A) are shown according to their position in the genome map at the top of panel B. The passage (P) in which these mutations were identified is indicated to the left of each genome map line. The amino acid mutations are color-coded according to the ORF in which they were found (colors described above). As indicated in the boxed legend, which applies to both panels, cyclophilin A binding loop mutations that have been previously identified as tissue culture adaptations are marked with a dagger symbol, and previously described nelfinavir-specific mutations are marked with an asterisk (with all references for these in the text and Tables 1 and 2).