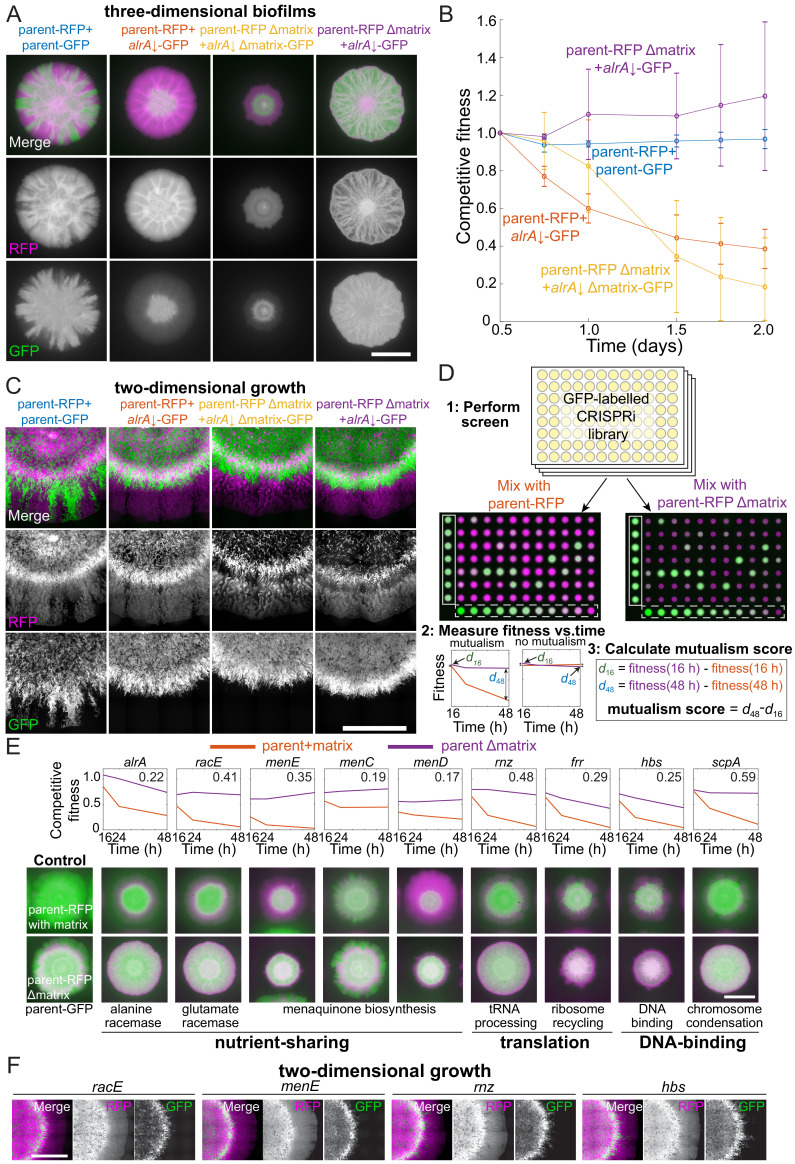
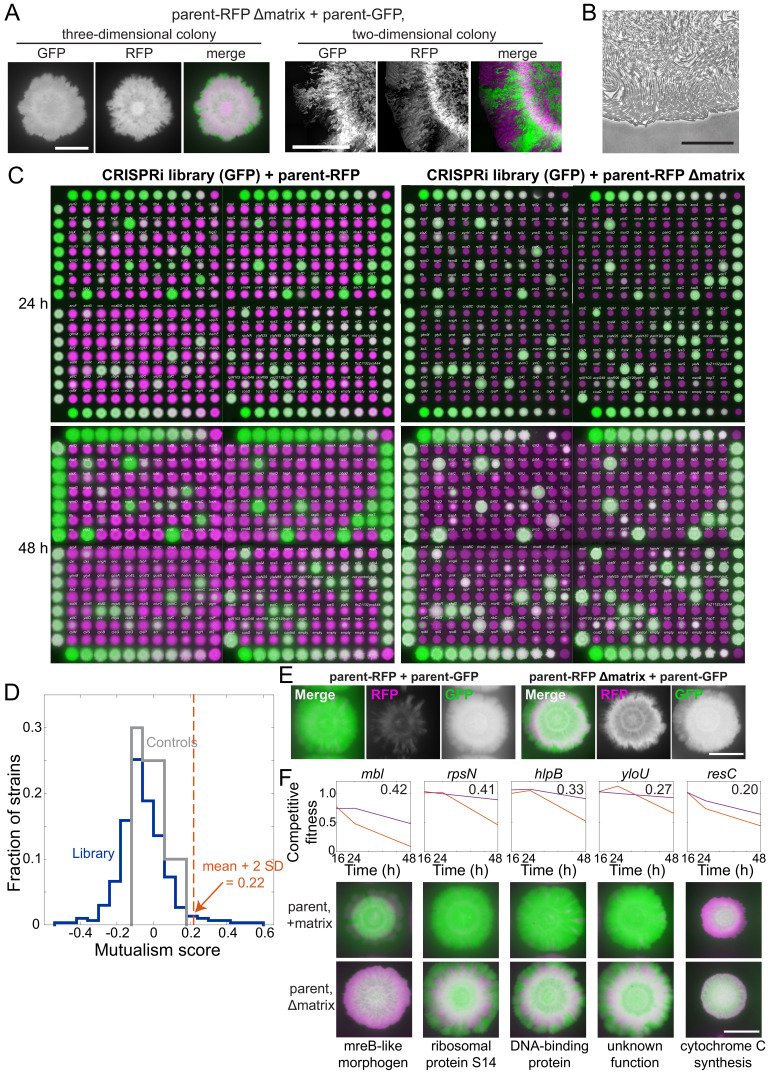
Figure 5. Mutualisms emerge when a nutrient-deficient mutant is the sole provider of extracellular matrix.
(A) Extracellular matrix is not required for alrA rescue (third column), and rescue is enhanced when the alrA full knockdown is combined with a matrix-deficient parent strain (fourth column). In the left column, the parent-GFP strain is false-colored in green, and in the other columns the alrA knockdown strain is green. The parent-RFP strain is false-colored in magenta. In the first two columns, both strains express matrix proteins. In the third column, both strains lack epsH and tasA, which encode key matrix components. In the fourth column, epsH and tasA are deleted from the parent-RFP strain while the alrA knockdown produces matrix. Images were acquired after 48 hr of growth. Scale bar: 5 mm. (B) Competitive fitness of the alrA full knockdown decreased over time when both strains or neither produced matrix, while fitness remained 1 (equal proportions of the two strains) and stable over time in co-cultures when the parent was matrix-deficient. The matrix-deficient parent-RFP+alrA knockdown (purple) and the parent-RFP+parent-GFP control (blue) data were not significantly different (p>0.2 at all time points); nor were the parent-RFP+alrA knockdown (orange) and the matrix-deficient parent-RFP+matrix-deficient alrA knockdown (yellow) data (p>0.07, Student’s t-test). The matrix-deficient parent-RFP+alrA knockdown (purple) fitness data were significantly higher than the parent-RFP+alrA knockdown (orange) at all time points after 0.5 days (p<0.014). Data were normalized to fitness at 0.5 days. Curves are means, and error bars represent one standard deviation (n = 2–3 biological replicates). Statistical analysis: Student’s unpaired t-test with a Benjamini–Hochberg multiple-hypothesis correction. (C) Cells with full knockdown of alrA were outcompeted by the parent-RFP strain during two-dimensional growth. Co-cultures were spotted on a MSgg-xylose agar pad and limited to growth in a layer with thickness one cell by applying a cover slip over the cells. Images were acquired after 24 hr of growth. Scale bar: 0.4 mm. (D) Design of screen to identify mutants that exhibit an increase in fitness when co-cultured with a matrix-deficient parent on MSgg-xylose agar. The sacA::gfp library was used in this screen; plate 3 from the 16 hr time point is shown. The distance between the centers of each colony is 9 mm. White box, controls; white dashed box, titration row. (E) Full knockdowns that exhibited mutualism generally involve genes related to nutrient sharing, translation, and DNA binding. Top: competitive fitness of co-cultures with the wild-type parent-RFP (purple) and matrix production-deficient parent (red) at 16, 24, and 48 hr. Numbers in the top right indicate the mutualism score. Bottom: merged images of biofilm colony co-cultures on MSgg-xylose agar at 48 hr. Scale bar: 5 mm. (F) Full knockdowns that exhibited mutualism in three-dimensional biofilm colonies were outcompeted by the matrix-deficient parent-RFP strain when growth was limited to two dimensions as in C. Images were acquired after 24 hr of growth. Scale bar: 0.4 mm. For (A, C–F), GFP is false-colored in green and RFP is false-colored in magenta.