The varying influenza A virus (IAV) exposure and infection status of individual swine facilitates introduction, transmission, and dissemination of diverse IAVs. Since agricultural fairs bring people into intimate contact with swine, they provide a unique interface for zoonotic transmission of IAV. Understanding the dynamics of IAV transmission through exhibition swine is critical to mitigating the high incidence of variant IAV cases reported in association with agricultural fairs. We used genomic sequences from our exhibition swine surveillance to characterize the hemagglutinin and full genotypic diversity of IAV at early-season shows and the subsequent dissemination through later-season agricultural fairs. We were able to identify a critical time point with important implications for downstream IAV and zoonotic transmission. With improved understanding of evolutionary origins of zoonotic IAV, we can inform public health mitigation strategies to ultimately reduce zoonotic IAV transmission and risk of pandemic IAV emergence.
KEYWORDS: genomic epidemiology, influenza A virus, public health, swine, zoonoses
ABSTRACT
Influenza pandemics are associated with severe morbidity, mortality, and social and economic disruption. Every summer in the United States, youths attending agricultural fairs are exposed to genetically diverse influenza A viruses (IAVs) circulating in exhibition swine, resulting in over 450 lab-confirmed zoonotic infections since 2010. Exhibition swine represent a small, defined population (∼1.5% of the U.S. herd), presenting a realistic opportunity to mitigate a pandemic threat by reducing IAV transmission in the animals themselves. Through intensive surveillance and genetic sequencing of IAVs in exhibition swine in six U.S. states in 2018 (n = 212), we characterized how a heterogeneous circuit of swine shows, comprising fairs with different sizes and geographic coverage, facilitates IAV transmission among exhibition swine and into humans. Specifically, we identified the role of an early-season national show in the propagation and spatial dissemination of a specific virus (H1δ-2) that becomes dominant among exhibition swine and is associated with the majority of zoonotic infections in 2018. These findings suggest that a highly targeted mitigation strategy, such as postponing swine shows for 1 to 2 weeks following the early-season national show, could potentially reduce IAV transmission in exhibition swine and spillover into humans, and this merits further study.
IMPORTANCE The varying influenza A virus (IAV) exposure and infection status of individual swine facilitates introduction, transmission, and dissemination of diverse IAVs. Since agricultural fairs bring people into intimate contact with swine, they provide a unique interface for zoonotic transmission of IAV. Understanding the dynamics of IAV transmission through exhibition swine is critical to mitigating the high incidence of variant IAV cases reported in association with agricultural fairs. We used genomic sequences from our exhibition swine surveillance to characterize the hemagglutinin and full genotypic diversity of IAV at early-season shows and the subsequent dissemination through later-season agricultural fairs. We were able to identify a critical time point with important implications for downstream IAV and zoonotic transmission. With improved understanding of evolutionary origins of zoonotic IAV, we can inform public health mitigation strategies to ultimately reduce zoonotic IAV transmission and risk of pandemic IAV emergence.
INTRODUCTION
The rapid evolution of influenza A viruses (IAVs) presents an ongoing threat to global health and economic security. IAVs have the capacity to host-switch into humans from animal reservoirs, particularly from avian and swine hosts (1), igniting global pandemics associated with severe morbidity, mortality, and social and economic disruption (2). Asia is considered a major source of emerging viruses with pandemic potential, owing to the high exposure of humans to diverse IAVs circulating in the multitude of domestic and wild species sold at live-animal markets (3). However, the evolutionary origins of the 2009 H1N1 pandemic in swine in Mexico (4) underscored the potential for novel pandemic viruses to emerge from North America and other regions and highlighted gaps in our understanding of the settings in which pandemic viruses emerge.
The evolution of pandemic viruses requires a “perfect ecological storm” in which multiple host populations perform at least two key functions: (i) generating and sustaining viral diversity and (ii) transmitting viral diversity to humans. Reservoir hosts serve the former purpose, harboring high viral genetic diversity but with low rates of direct contact with humans (such as wild birds, bats, and rodents) (5). Intermediary hosts are typically livestock or domestic animals that serve the latter purpose: they often harbor only a subset of the viral diversity that circulates in reservoir hosts yet are capable of transmitting viral diversity to humans via high rates of contact (1). In the United States, commercial swine serve as reservoir hosts, and exhibition swine effectively serve as intermediary hosts. Commercial swine are produced for food production exclusively in a year-round system that is highly conducive to the evolution and sustained transmission of genetically diverse viruses (6) but less conducive to zoonotic transmission, with high biosecurity and less frequent contact with humans. In contrast, exhibition swine effectively serve as intermediary hosts because they are raised and exhibited in a system with low biosecurity that maximizes human contact (7, 8) but purges genetic diversity and host immunity at the end of the summer show season, when exhibition swine finish showing and a new cohort of swine are raised for the next year (9). A production system in which commercial and exhibition swine are occasionally raised together provides opportunities for genetically diverse viruses to end up at county fairs and find their way into humans (10–12).
In the United States, zoonotic infections with swine-origin IAVs occur every summer at swine exhibitions (Fig. 1A). Although major outbreaks associated with hundreds of zoonotic infections have not been observed since 2012, the repeated infection of humans each year with increasingly genetically diverse IAVs presents an ongoing pandemic risk. Potential limited human-to-human transmission has been observed (13), and each infection of a human with a genetically different swine-origin virus increases the probability that a particular variant will acquire the capacity for sustained transmission in humans. The potential targets for reducing the risk of zoonotic infection at swine exhibitions include multiple interfaces for IAV transmission: (i) swine to human, at exhibitions; (ii) swine to swine, at exhibitions; (iii) show to show, via individuals that bring exhibition swine to multiple fairs in a summer; and (iv) commercial to exhibition swine, which introduces new IAVs into exhibition swine each summer.
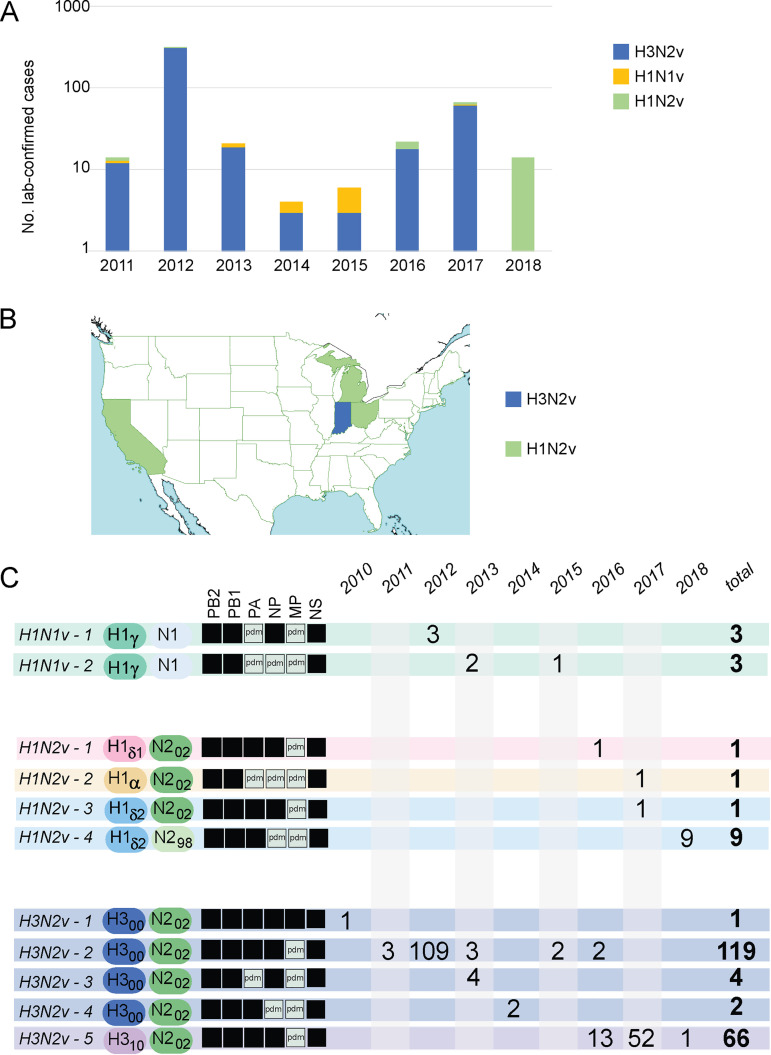
FIG 1.
Human infections with variant IAVs of swine origin in the United States, 2011 to 2018. A total of 463 human infections with variant IAVs were lab confirmed in the United States during 2011 to 2018. (A) The number of human infections that are associated with IAV subtypes in a given year is indicated by the height of the bar, on a log scale. (B) In 2018, human infections with variant IAVs were lab confirmed in four U.S. states: California (n = 6), Indiana (n = 1), Michigan (n = 3), and Ohio (n = 4). Data were downloaded from the CDC’s FluView interactive site. (C) The number of variant cases associated with 1 of 11 H1N1v, H1N2v, or H3N2v genotypes is indicated for each year, based on whole-genome sequence data available from GISAID.
The swine show circuit in the United States is a complex network composed of thousands of events that differ in size, geographic coverage of participants, and degrees of connectivity (Table 1). The vast majority of America’s 3,000+ counties hold a county fair every summer, and each of the 50 states holds a state fair, which occur throughout the year. Most agricultural fairs include a swine competition for youths ages 9 to 21. In addition to fairs, swine are also exhibited by youth ages 3 to 21 at jackpot shows, which are events held specifically to show swine and do not include the additional community festival attractions that are present at a typical fair. The ubiquity of swine shows means that the risk for zoonotic infections spans the entire United States, as reflected by the detection of zoonotic swine-origin variant IAVs (H1N1v, H1N2v, and H3N2v) in 22 geographically dispersed states, from New Jersey on the east coast to California on the west coast, and even in Hawaii (data available from the CDC FluView website [https://www.cdc.gov/flu/weekly/fluviewinteractive.htm]). However, the risk of zoonoses is not evenly distributed across the country; Ohio and Indiana account for over half of all swine-origin zoonoses since 2011. The shift in zoonotic infections from predominantly H3N2v during 2011 to 2017 to H1N2v in 2018 (13) raises questions about the degree to which the human-swine interface is deterministic, where certain IAV subtypes, genotypes, or mutations increase a virus’s intrinsic capacity to infect humans. Alternatively, in a stochastic model, the viruses associated with zoonotic infections in a given year have no specific characteristics conducive to human infection but randomly happen to be dominant in exhibition swine that season.
TABLE 1.
IAV surveillance in four types of U.S. swine shows
| Parameter | Result for show type |
||||
|---|---|---|---|---|---|
| Local jackpot | National/regional jackpot | State fair | County fair | All exhibitions | |
| Characteristics of shows | |||||
| Geographical restrictions | None | None | State | County | |
| Size (no. of pigs) | Small/medium | Large | Large | Small/medium | |
| Duration (days) | Short | Long | Long | Long | |
| Timing | Early summer | All yr | Late summer | All summer | |
| Sampling strategy | |||||
| No. of shows/fairs sampled | 4 | 6 | 5 | 98 | 113 |
| Time of sampling (mo) | May–Jun | Jan–Dec | Jul–Aug | Jun–Oct | Jan–Dec |
| Location of sampling (states) | OH | AZ, GA, IA, IL, KY | IA, IN, KY, OH, WV | IN, MI, OH | AZ, GA, IA, IL, IN, KY, MI, OH, WV |
| Total no. of samples collected | 594 | 2,106 | 1,012 | 1,992 | 5,704 |
| IAV detection | |||||
| PCR, % positive | |||||
| Shows | 100.0 | 100.0 | 80.0 | 42.9 | 49.6 |
| Samples in positive shows | 17.2 | 12.9 | 9.7 | 42.0 | 18.2 |
| Total samples | 17.2 | 12.9 | 9.5 | 17.9 | 14.5 |
| VI, % positive | |||||
| Shows | 100.0 | 83.3 | 80.0 | 32.7 | 39.8 |
| Samples in positive shows | 6.4 | 6.2 | 3.5 | 37.1 | 10.3 |
| Total samples | 6.2 | 6.4 | 3.5 | 12.1 | 7.8 |
To better understand the evolutionary and ecological dynamics of IAVs at swine exhibitions in 2018, we sequenced the entire genome of 212 IAVs collected from exhibition swine at 113 fairs in six U.S. states and performed a phylogenetic analysis, including additional samples from humans and commercial swine acquired from public databases. We found that the frequency of H1N2v zoonoses in 2018 is likely to be stochastic, arising from the high frequency of H1N2 (H1δ-2) viruses in exhibition swine that year. Furthermore, we traced the high frequency of H1δ-2 viruses in exhibition swine that year to a specific amplification event that occurred at an early-season national jackpot show, which facilitated the coast-to-coast dissemination of H1δ-2 viruses. These findings have important implications for understanding how the structure of the U.S. swine show circuit facilitates IAV transmission in exhibition swine and zoonoses.
RESULTS
IAV surveillance in U.S. swine shows in 2018.
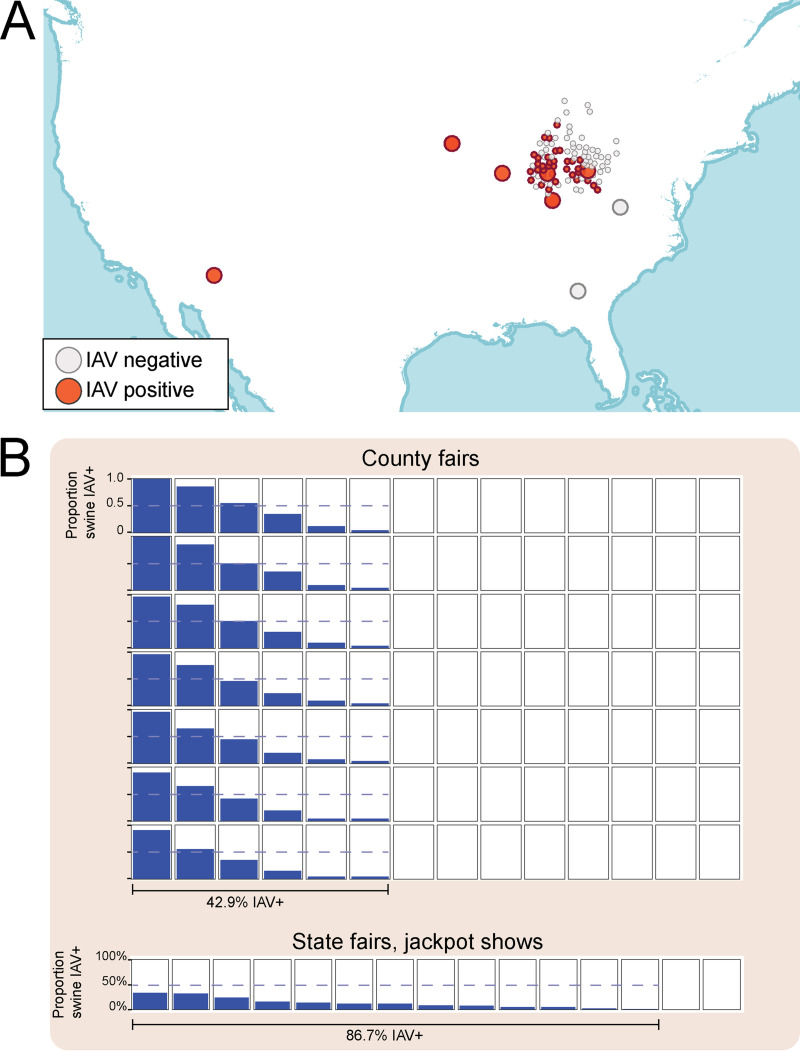
From January to December 2018, 5,704 samples were collected from swine at 113 exhibitions in nine U.S. states (Table 1). IAV-positive shows were identified in seven of the nine U.S. states sampled for this study (Fig. 2A). Overall, almost 40% of shows were positive for IAV by virus isolation (VI) (Table 1). Approximately one-third of county fairs were IAV positive by VI, whereas 90% of jackpot shows and 80% of state fairs were IAV positive. However, when IAV was detected at a county fair, a larger proportion of pigs tested positive (42% by PCR [mean]), compared to state fairs, local jackpot shows, or national jackpot shows (10%, 13%, and 17%, respectively, by PCR [means]) (Table 1; Fig. 2B). Across all shows, IAV was isolated from PCR-positive samples with 54% VI efficiency (Table 1; Fig. 2B).
FIG 2.
IAV surveillance in U.S. exhibition swine in 2018. (A) Each circle represents one of the 113 swine exhibitions from which respiratory samples obtained from pigs and tested for IAV. The color of the point indicates whether the exhibition was IAV positive (red) or negative (gray) by virus isolation. Large points represent larger shows (state fairs and national or regional jackpot shows); small points represent smaller shows (local jackpot or county fairs). (B) Each box represents a swine show that was sampled for this study, including county fairs (top; n = 98) and state fairs and jackpot shows (bottom; n = 15). Within each box, the blue shading indicates the proportion of swine that tested positive for IAV by PCR at that individual show. The horizontal dotted blue line indicates 50% of pigs testing positive. A single line below indicates the proportion of all shows of a certain type (county or state/jackpot) that had at least one pig test positive for IAV.
Genetic diversity of HA in U.S. exhibition swine in 2018.
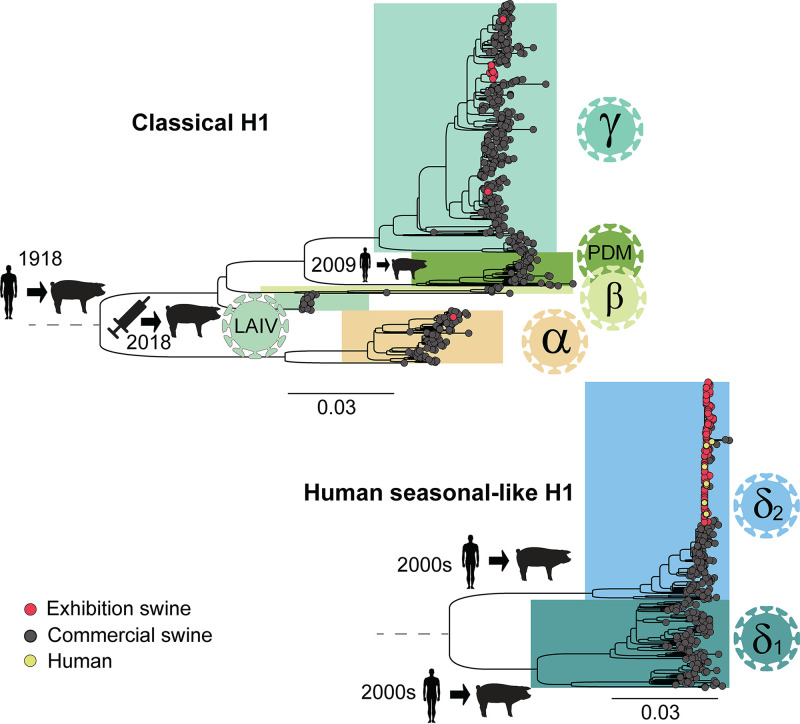
Five genetically distinct HA lineages were identified in U.S. exhibition swine in 2018: H1α (1A.1), H1γ (1A.3), H1δ-2 (1B.2.1), H3 (2000s), and H3 (2010s) (Fig. 3; Fig. S1). Seven additional HA lineages were observed in U.S. commercial swine in 2018 that were not observed in exhibition swine, including four H1 lineages (H1β, H1N1pdm09, LAIV [live attenuated influenza vaccine]-like H1 viruses, and H1δ-1) (Fig. 3) and three H3 lineages (H3 (2016), H3 (2017) and LAIV-like H3) (Fig. S1). However, H1δ-1 (1B.2.2) viruses were the only viruses widely circulating in U.S. commercial swine in 2018 that were not observed in exhibition swine in 2018.
FIG 3.
Phylogenetic relationships of H1 viruses circulating in U.S. swine in 2018. Maximum-likelihood trees inferred for H1 segments (n = 984) of IAVs collected in the United States during 2018. At the tips of the tree, red circles indicate viruses collected for this study from U.S. exhibition swine (n = 196), dark gray circles indicate background sequences downloaded from GenBank that are primarily from U.S. commercial swine (n = 779), and yellow circles indicate human infections with variant swine-origin viruses (H1N2v) downloaded from GISAID (n = 9). All horizontal branch lengths are drawn to scale (nucleotide substitutions per site). Trees are midpoint rooted for clarity, and bootstrap values of >70 are provided for key nodes. The top tree represents the classical (CswH1) branch, which descends from the 1918 Spanish influenza pandemic: (i) H1α (1A.1), (ii) H1β (1A.2), (iii) the currently dominant H1γ viruses (1A.3.3.3), (iv) the 2009 H1N1 pandemic virus (H1N1pdm09, 1A.3.3.2) that was introduced from humans into U.S. swine herds following the 2009 H1N1 pandemic, and (v) H1 viruses related to late 1990s vaccine strains contained in the live attenuated influenza vaccine (LAIV) that was licensed for use in U.S. swine in 2018. The bottom tree represents the human seasonal-like H1 viruses (H1δ-1, H1δ-2, 1B.2.2, and 1B.2.1 [37]) that were introduced from humans on two separate occasions during the early 2000s. Major introductions of IAVs from human into swine are indicated along respective branches.
Whole-genome diversity of IAV in U.S. exhibition swine.
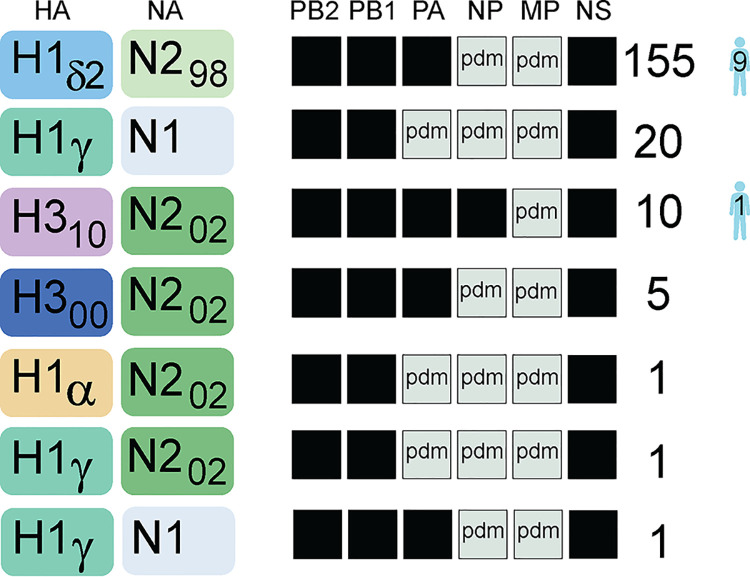
In addition to the genetic diversity of the HA, three genetically distinct NA lineages were identified in exhibition swine: N1 (classical), N2 (1998), and N2 (2002) (Fig. 4). All viruses found in exhibition swine had PB2, PB1, and NS segments from the TRIG (triple reassorted internal genes) lineage and MP from the 2009 H1N1pdm lineage (Fig. 4). The majority of PA segments (∼89%) derived from the TRIG lineage, and ∼95% of NP segments derived from the 2009 H1N1pdm lineage. Approximately 80% of the viruses for which complete genome sequences were available (155/193) had a genotype with an H1δ-2 hemagglutinin segment, representing the first time that H1δ-2 viruses have been dominant in 9 years of surveillance in U.S. exhibition swine (Fig. 1C). All H1δ-2 viruses had the same genetic background, including N2 (1998) and internal genes from the TRIG (PB2, PB1, PA, and NS) and 2009 H1N1pdm lineages (NP and MP). The remaining 20% of exhibition swine viruses had an HA segment from one of four lineages: H1α (n = 1), H1γ (n = 22), H3N2 (2000) (n = 5), and H3N2 (2010) (n = 10). Overall, seven reassortant genotypes were identified in U.S. exhibition swine in 2018 (Fig. 4). The genotypes observed in exhibition swine were similar to genotypes circulating in U.S. commercial swine at that time (Fig. S2), indicating that the genotype constellations were likely formed before being introduced from commercial swine into exhibition swine.
FIG 4.
Seven reassortant genotypes identified in U.S. exhibition swine during 2018. Each box represents one of the eight segments of the IAV genome. Antigenic segments (HA and NA) are listed first, followed by the six internal gene segments. The shade of the box corresponds to the genetic lineage of the segment. For HA: yellow, H1α; sea green, H1γ; light blue, H1δ-2; dark blue, H3N2 (2000s lineage); purple, H3N2 (2010s lineage). For NA: light blue, N1 (classical); light green, N2 (1998 lineage); dark green, N2 (2002 lineage). For internal genes, black indicates the TRIG lineage and light green indicates the 2009 H1N1pdm lineage. The number of viruses with a particular genotype that were identified in U.S. exhibition swine and humans in 2018 is indicated.
Whole-genome diversity of zoonotic viruses in humans.
In 2018, zoonotic infections were detected in six U.S. states: California, Colorado, Indiana, Iowa, Michigan, and Ohio (Fig. 1B). Following eight summers where the majority of zoonotic infections at U.S. swine shows were associated with the H3N2 subtype (H3N2v), the vast majority of zoonotic infections in 2018 were associated with H1N2v viruses (Fig. 1C). The nine H1N2v human cases for which whole-genome sequences were available had the H1δ-2 genotype, similar to what was observed most frequently in exhibition swine (Fig. 4). The virus in the one H3N2v human case had a genotype similar to the only genotype observed with a HA from the H3N2 (2010) lineage. The high frequency of H1N2v cases identified in 2018 represents a departure from the dominance of H3N2v cases during 2010 to 2017. Of the 210 variant viruses for which whole-genome sequence data are available, 91% are from the H3N2 subtype (Fig. 1C). However, a variety of genetically diverse viruses have been recovered from humans, including six different HA lineages (H1α, H1γ, H1δ-1, H1δ-2, H3 [2000s], and H3 [2010s]) and 11 different genotypes (Fig. 1C). The vast majority (99%) of variant viruses identified in humans have reassortant genotypes that emerged after the introduction of H1N1pdm internal gene segments into U.S. swine from humans following the 2009 pandemic.
Multiple introductions of IAV from U.S. commercial to exhibition swine during 2018.
Repeated introductions of genetically diverse IAVs from commercial swine appear to be the major source of viral diversity observed in exhibition swine in 2018. The phylogenetic tree inferred for IAV sequences collected from U.S. commercial and exhibition swine indicates that there were seven discrete introductions of IAVs from commercial swine into exhibition swine during 2018 (Fig. 5). Each of the five HA lineages (H1δ-2, H1α, H1γ, H3N2 [2000s lineage], and H3N2 [2010s lineage]) was introduced separately from commercial swine, and H1γ viruses were introduced two additional times (three H1γ introductions total). The frequency of an HA lineage being introduced into exhibition swine was proportional to its frequency in the U.S. commercial swine population. For example, H1γ viruses that are most frequently observed in commercial swine were introduced three times, whereas low-frequency HA lineages, such as H1pdm, were never introduced. H1δ-2, H1α, H1γ, H3 (2000), and H3 (2010) HA lineages, all with moderate detection in commercial swine, were introduced into exhibition swine once. H1δ1is the only HA lineage that was routinely detected in U.S. commercial swine during the first 5 months of 2018 (∼15% of viruses) but was never detected that year in exhibition swine. However, high frequency of an HA lineage in U.S. commercial swine was not associated with detection at a larger number of swine shows.
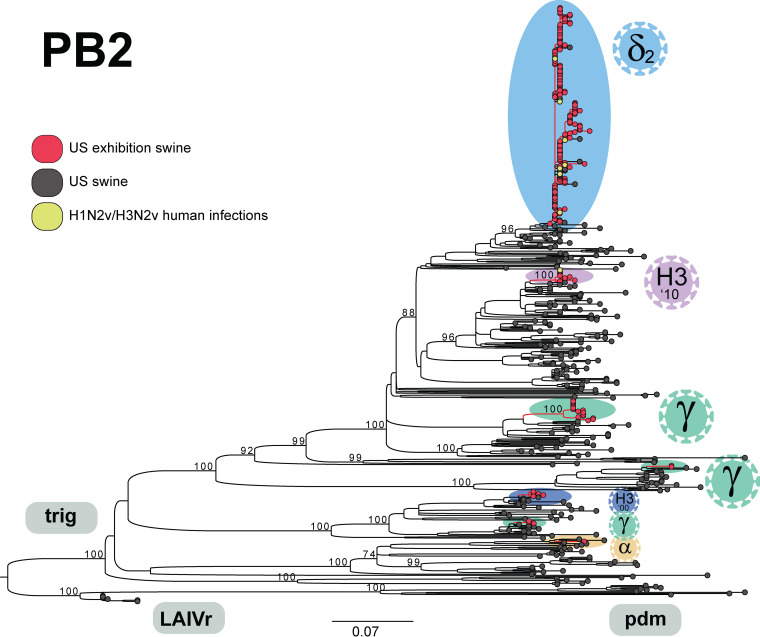
FIG 5.
Phylogenetic relationships of IAVs collected in U.S. swine during 2018. Maximum-likelihood tree inferred for PB2 segments (n = 526) of IAVs collected in the United States during 2018. Viruses collected for this study from U.S. exhibition swine are shaded red (n = 212); background sequences downloaded from GenBank are primarily from U.S. commercial swine and shaded black (n = 304); human infections with variant swine-origin viruses (H1N2v or H3N2v) were downloaded from GISAID and are shaded yellow (n = 10). All horizontal branch lengths are drawn to scale (nucleotide substitutions per site). The tree is midpoint rooted for clarity, and bootstrap values of >70 are provided for key nodes. Seven discrete introductions of IAVs into exhibition swine during 2018 are labeled by HA lineage (shading similar to that in Fig. 1).
Spatial dissemination of H1δ-2 viruses.
An important observation is that, although four antigenically distinct viruses were circulating during the early phase of the 2018 exhibition swine show season (H1γ, H1δ-2, H3 [2000], and H3 [2010]), only H1δ-2 viruses were transmitted onward at high frequencies and were detected repeatedly in July and August at the county and state fairs, where the majority of zoonotic transmission events occur (Fig. 6). H1γ viruses, despite being introduced multiple times from commercial swine, presented relatively little evidence of onward transmission among exhibition swine, with detection at only 1.67 shows, on average, per introduction. In contrast, the single introduction of H1δ-2 viruses was detected at 38 shows spanning six U.S. states: Kentucky, Illinois, Iowa, Indiana, Michigan, and Ohio. H1δ-2 viruses were also detected at all four types of swine shows (local jackpot shows, national jackpot shows, county fairs, and state fairs). In Ohio, for example, H1δ-2 viruses were detected at multiple local jackpot shows and county fairs, as well as the state fair, prior to detection in humans in August (H1N2v). Human H1N2v cases were primarily observed in states where H1δ-2 viruses had previously been identified in exhibition swine, with the exception of California, in which swine were not sampled for this study. Excluding California, we estimate that H1δ-2 viruses disseminated at least 2,576 km in exhibition swine, a lowest-bound estimate based on the shortest route inferred between all 38 swine shows where H1δ-2 viruses were detected and not matching the actual timing of shows.
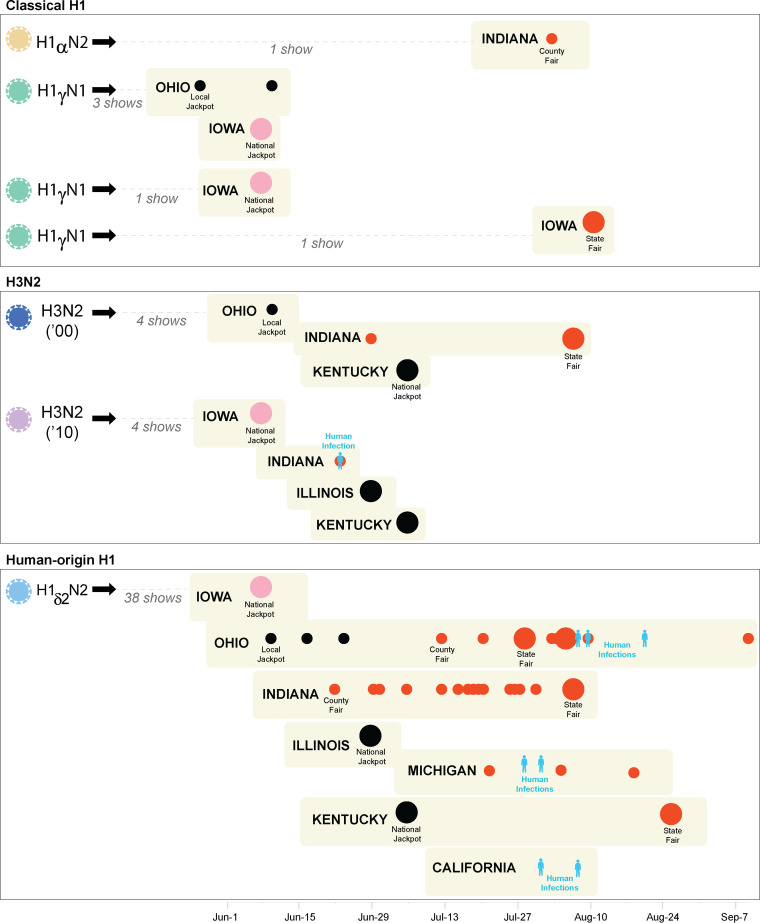
FIG 6.
Seven introductions of IAV into U.S. exhibition swine during 2018. Each black arrow indicates a discrete introduction of IAV into U.S. exhibition swine.The color of the cartoon virus represents the subtype and lineage of the virus: yellow, H1α; green, H1γ; light blue, H1δ-2; dark blue, H3N2 (2000s); and purple, H3N2 (2010s). Swine shows where genetically related viruses associated with each introduction were observed are indicated, separated vertically by state and ordered along the x axis by the date of virus sampling at the fair. Large pink circles indicate early-season national jackpot shows in Iowa, large black circles indicate other national jackpot shows, small black circles indicate local jackpot shows, small red circles indicate county fairs, and large red circles indicate state fairs. Human infections are indicated in blue. Shows are organized vertically by introduction and U.S. state. The total numbers of shows where an introduction was identified are given.
IAV diversity at an early-season national jackpot.
Intensive surveillance of an early-season national jackpot show (57 whole-genome sequences) (14) provided a detailed picture of the viral diversity circulating at the start of the show season at an exhibition that draws pigs from across the United States. Thirteen of these IAVs were previously described in a study demonstrating the utility of portable nanopore sequencing platforms for real-time surveillance at swine shows (14). Four independent introductions of IAVs were observed at the show, including two introductions of H1γ viruses, one introduction of H3N2 (2010) viruses, and one introduction of H1δ-2 viruses (Fig. 6). However, only H1δ-2 viruses had evidence of widespread transmission between pigs attending the show, constituting more than 80% (47/57) of the IAVs sequenced from this national show (Fig. 7). Using zip code data from exhibitors, we determined that H1δ-2 viruses infected exhibition swine that had been transported to the national show from 17 U.S. states spanning over 4,800 kilometers, providing opportunities for long-distance viral dissemination when the pigs returned to their home states (Fig. 8). For example, H1δ-2 viruses were identified in four swine from California that attended the national jackpot show (Fig. 8). Although the viruses are genetically similar to viruses associated with the zoonotic infections in California, the national jackpot show occurred 2 months before the zoonotic infections. It is likely that H1δ-2 viruses were introduced multiple times from the Midwest into California, but the lack of IAV sequence data from exhibition swine in California makes it impossible to prove or disprove this hypothesis.
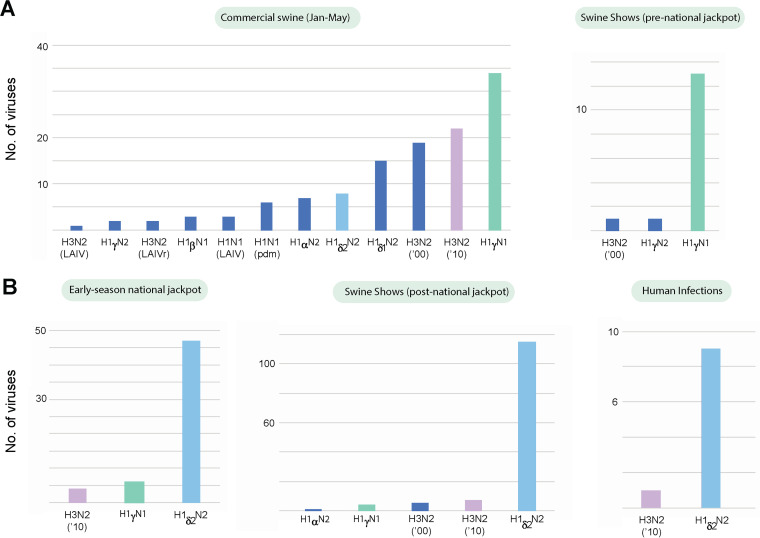
FIG 7.
Distribution of IAV subtypes, before and after the national jackpot. Prior to the early-season national jackpot show, H1γ viruses are most frequently observed in commercial and exhibition swine (A). H1δ-2 viruses are most frequently observed during the early-season national jackpot show, at all subsequent swine shows (June to September), and in humans (B).
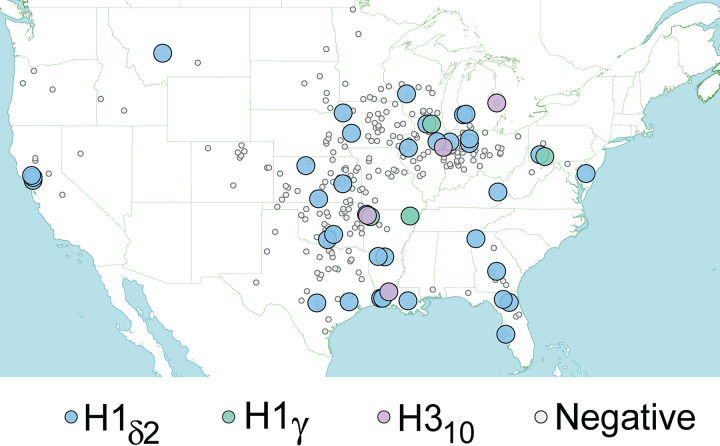
FIG 8.
Spatial distribution of exhibition swine infected with IAVs at an early-season national jackpot show. Each circle represents the home farm of a pig that attended the 2018 early-season national jackpot show and was tested for IAV. Small gray circles indicate pigs that tested negative for IAV. Large circles indicate pigs that tested positive for IAV, with the shade of the circle corresponding to the specific HA lineage.
Critically, the H1δ-2 viruses that were dominant at the national show continued to be dominant at swine shows for the remainder of the summer and were subsequently isolated at 92% (35/38) of the season’s remaining VI-positive swine exhibitions (Fig. 7). Ninety percent of zoonotic infections in humans that occurred in late July and August also were associated with the single clade of H1δ-2 viruses that can be traced back to the early-season national show (Fig. 7). Overall, 94.7% (142/150) of viruses sequenced from shows following the early-season national jackpot show were genetically related to viruses identified at the national show. Only two shows (5.3%) had viruses that were genetically distinct from those at the national show, representing late-season introductions of H1α and H1γ viruses (Fig. 6).
Emergence and evolution of H1δ-2 viruses in U.S. swine, 2005 to 2019.
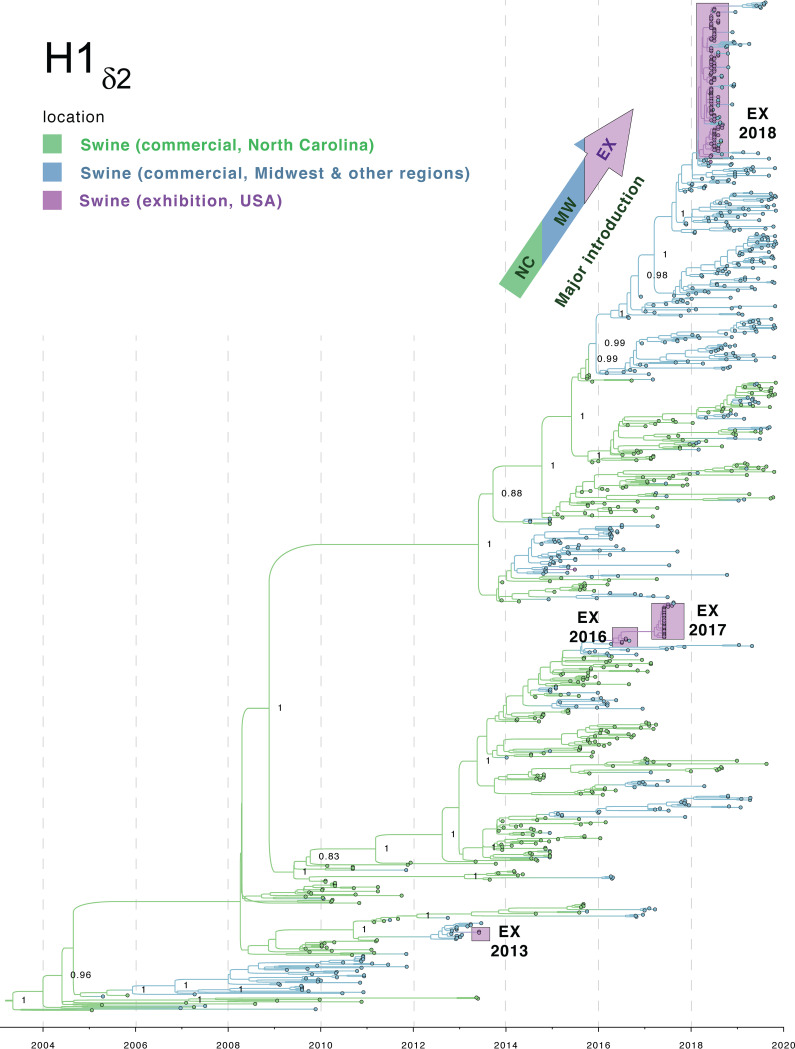
To better understand how evolutionary events in commercial swine led to the emergence and dominance of H1δ-2 viruses in exhibition swine in 2018, and the resulting spillover infections of humans, a time-scaled maximum clade credibility (MCC) tree was inferred for the 950 H1δ-2 HA sequences available from our study and in GenBank since 2005 (Fig. 9). The tree is consistent with prior estimates that H1δ-2 viruses originated in commercial swine in North Carolina, where they were introduced from humans in the early 2000s. These viruses have disseminated dozens of times to other states, primarily in the Midwest region, following commercial “swineways” (15). While the H1δ-2 virus has circulated for over 15 years in commercial swine in North Carolina, the vast majority of introductions into Midwest swine died out after only a few years of circulation. Before disappearing, several of the H1δ-2 introductions into the Midwest commercial swine population did manage to transmit to exhibition swine in 2013, 2015, 2016, and 2017 (Fig. 9). Each time, however, the H1δ-2 viruses experienced only limited transmission in exhibition swine and from 2011 to 2018 were associated with only one zoonotic infection (Fig. 1C).
FIG 9.
Phylogenetic relationships of H1δ-2 viruses collected in U.S. swine, 2005 to 2019. Time-scaled MCC tree inferred for 950 HA (H1δ-2) sequencesfrom IAVs collected in U.S. swine since 2005. The color of each branch indicates the most probable location state: green, commercial swine in North Carolina; blue, commercial swine from other U.S. states, primarily in the Midwest region; purple, exhibition swine from all U.S. states collected for this study. Posterior probabilities are provided for key nodes. The four introductions of H1δ-2 viruses into exhibition swine are indicated by purple boxes. The arrow at the top of the tree indicates the major introduction of H1δ-2 viruses from North Carolina into the Midwest region and thereafter into exhibition swine in 2018. The origins of H1δ-2 viruses via a human-to-swine transmission event in the early 2000s is indicated near the root of the tree.
The H1δ-2 viruses identified in exhibition swine in 2018 belong to a new clade of H1δ-2 that has become established in Midwest commercial swine herds in recent years, reaching Iowa and Illinois by the end of 2016 and Minnesota, Missouri, and Nebraska by June 2018. The clade is associated with a new genotype: HA (H1δ-2), N2 (1998), PB2 (TRIG), PB1 (TRIG), PA (TRIG), NP (pdm), MP (pdm), and NS (TRIG) (Fig. 4). This genotype is similar to the previously dominant H1δ-2 genotype in all segments except the NP segment, which changed from TRIG to pdm following a reassortment event that occurred during 2014-2015, just before the clade was introduced from North Carolina to the Midwest (Fig. S3). The success of this clade has led to an increase in the proportion of H1 viruses that belong to the H1δ-2 lineage in the U.S. states of Iowa, Indiana, Illinois, Ohio, and Missouri (corn belt region) as well as Minnesota, Wisconsin, and Michigan (Great Lakes region). The proportion has increased from <5% in 2014 to >30% between 2014 and 2019, approaching the levels observed in North Carolina where, since 2005, the 15-year average proportion of H1δ-2 IAVs identified exceeds 30%. It is possible that the widespread dissemination of H1δ-2 viruses in exhibition swine in 2018 also contributed to further spatial spread of H1δ-2 viruses in commercial swine, as periodic exhibition-to-commercial swine transmission also has been documented. Viruses collected from commercial swine in western and southern states, including Arkansas, Florida, Oregon, and California, are dispersed within the large clade of H1δ-2 viruses collected from exhibition swine in 2018 (Fig. 9).
DISCUSSION
By further characterizing IAV circulation in the swine show circuit in the United States, our study’s findings identify potential strategies for reducing zoonotic transmission of IAVs. Zoonotic IAV infections recur each year when large numbers of swine exhibited at county and state fairs become infected with IAV. Our study traced the evolutionary origins of zoonotic viruses back through a complex network of swine shows in which animals with different travel and exposure histories interact with each other and with humans. To date, jackpot shows have not been a focus of IAV surveillance, since they have not been directly associated with zoonotic infections. However, jackpot shows occur prior to county fairs and serve as centers for intermingling between pigs transported from distant locations. Our study suggests that an early-season national jackpot show in 2018 may have had an important role in the emergence and long-distance dissemination of the H1δ-2 viruses that traveled over 2,500 km and later appeared at numerous county fairs and in humans. In fact, it is possible that hundreds of pigs at this one show were infected with the H1δ-2 virus, given that 3,000 swine participated, approximately one-third of randomly sampled animals were IAV-positive by PCR, and >80% of viruses sequenced from the show belonged to the H1δ-2 lineage. These findings suggest that targeted strategies to reduce zoonoses and IAV transmission at U.S. swine shows will likely require a greater understanding of the role of jackpot shows in seeding outbreaks. However, the sheer number of events, animals, and interfaces introduce considerable stochasticity into the transmission network, which complicates mitigation and prediction. The outsized effect of one early-season national jackpot show on IAV transmission in 2018 could be an anomaly, and further study is needed to determine whether patterns occur consistently across years.
The fact that the zoonotic infections occurred at least 6 weeks after the early-season national show highlights how zoonotic risk is generated not by a single event but by a vast interconnected network of exhibitions. The inclusion of frequent local shows as well as sporadic national shows creates a particularly robust network capable of both sustaining IAV transmission over the duration of the show season and disseminating viruses over large geographical areas. The robustness of the network is also maintained by the participation of a small number of potential “superspreading” pigs that attend 20 or more shows each year and travel long distances for competitions. The majority of pigs attend relatively few shows (for local-fair exhibitors, x̅ = 2 shows/year; for jackpot exhibitors, x̅ =11 shows/year) and are exposed to fewer IAVs (16, 17). The presence of large numbers of immunologically naive animals likely contributes to rapid within-show transmission.
It is important to understand how the swine show network is embedded within a larger system of human and swine populations that play different roles in IAV transmission. In general, the flow of IAVs occurs primarily in a single direction: (i) commercial swine primarily acquire viruses from humans, particularly swine workers; (ii) exhibition swine acquire viruses from commercial swine; and (iii) youth exhibitors acquire zoonotic infections from exhibition swine. From a public health perspective, we naturally focus on swine as sources of variant viruses for humans. But it is important to keep in mind that most of the IAVs that circulate in commercial swine are of human origin, including the H1δ-2 viruses introduced from humans into U.S. commercial swine in the early 2000s (18).
Youth exhibitors and fairgoers are exposed each summer to a different subset of the genetically diverse IAVs that are introduced from U.S. commercial swine into exhibition swine. The reasons for the higher rates of detection of zoonotic IAVs infections at swine exhibitions in the United States beginning in 2011 remain unclear. There may be biological explanations, as the vast majority of zoonotic viruses are reassortants that emerged following the introduction of the 2009 H1N1 pandemic virus into U.S. swine. At the same time, recognition of the zoonotic risk of IAVs in swine greatly increased following the 2009 pandemic, and it is possible that the uptick in zoonotic infections is attributable to increased public health awareness and detection. Even so, there are many limitations to the detection of zoonotic IAV infections during summer months, when physicians in the United States do not typically test for influenza.
To date, interventions aimed at reducing zoonotic transmission at agricultural fairs have encountered numerous barriers. Behavioral changes encouraged by education and handwashing stations positioned outside swine barns are difficult to sustain when the perceived risk for zoonoses is low (19). Although exhibition swine are frequently infected with IAVs (20–22), it is difficult to perceive disease risk in sources considered as wholesome as county fairs or in exhibition swine that are groomed and treated like pets. Numerous mitigation strategies have been suggested, but each is not without limitations. Portable IAV sequencing and analysis platforms have provided real-time IAV data crucial to evaluating the risk of viral transmission and zoonoses (14). Moving forward, those types of data could inform rapid proactive intervention. However, event schedules and procedures can be difficult to alter, and whether such information will prove actionable remains unclear.
It has been proposed that swine exhibitors could be required to present documentation that their pigs had received an IAV vaccination to participate in a show (23, 24). However, IAV vaccine usage in exhibition swine already appears to be high, with 62% of county fair exhibitors and 71% of jackpot swine exhibitors reporting that their pigs had been vaccinated against IAV (16, 17). Although market shares of commercially available vaccines are unknown, interestingly, a commercially licensed IAV vaccine for swine had the H1δ-2 strain removed in the 2016 vaccine strain update (https://www.zoetisus.com/products/pork/flusure-xp/flusurexp.aspx). It is, however, difficult to speculate whether the observed increase of H1δ-2 IAVs in exhibition swine is associated with exclusion from commercially available IAV vaccines. It is difficult to ascertain what vaccines swine exhibitors are using, since shows with vaccine mandates often do not specify which type or brand of vaccination is to be administered. A better evaluation of exhibitor compliance could provide better insights into the benefits and best implementation strategies of IAV vaccination in this population of swine. Upstream efforts to prevent IAVs from transmitting from commercial to exhibition swine in the first place would be optimal, but little is known about operations that jointly raise commercial with show pigs and the commercial-exhibition interface. The fact that the swine show circuit is publicly advertised and well documented may provide the most realistic opportunity for identifying key events with outsized impacts on transmission across the entire network. While IAV surveillance at U.S. swine shows has focused on state and county fairs where zoonotic transmission typically occurs, our study suggests that national jackpot shows may present an earlier intervention point that warrants further investigation.
The emergence of a new clade of reassortant H1δ-2 viruses in both commercial and exhibition swine in the Midwest also warrants further investigation. The genetic composition of IAVs circulating in commercial swine in the Midwest each year has a major impact on which viruses spill over into exhibition swine and eventually humans. Since 2010, humans in the United States have been infected with at least 11 IAV genotypes with six different HAs. Gaps in our understanding of how and where viruses transfer from commercial to exhibition swine each year, combined with stochastic effects, make it difficult to predict which IAVs become predominant in exhibition swine each year. In general, it is not surprising that emerging viruses in Midwest commercial swine over the last several years would eventually spill over into exhibition swine. In addition, novel reassortant H1α viruses that recently spread from Canada into the Midwest (25) were also detected in exhibition swine in 2018, along with a novel H3N2 virus identified in 2015 that was introduced from humans (26). In future studies, it would be useful to better quantify how the dynamics of larger populations of viruses circulating in commercial swine in the Midwest (or nationwide) relate to the subset of viruses that are detected each year in exhibition swine. Our ability to predict which viruses are most likely to spill over from commercial to exhibition swine is impeded by biases and gaps in IAV surveillance in commercial swine herds, particularly in states with large exhibition swine populations, such as Ohio. The USDA conducts year-long IAV surveillance in U.S. herds, but the approach is passive and subject to biases in submission rates by location and producer (27). Given the limited resources available for studying IAV in swine, particularly compared to humans and avian species, an efficient approach to population-level surveillance in commercial herds may require the identification of specific production systems that could serve as regional sentinels (28).
MATERIALS AND METHODS
Active IAV surveillance in U.S. exhibition swine.
For this study, active surveillance of IAVs in exhibition swine was conducted during January to December 2018 at 113 swine shows located in nine U.S. states: Arizona, Georgia, Illinois, Indiana, Iowa, Kentucky, Michigan, Ohio, and West Virginia (Table 1). Surveillance included four types of swine shows that varied in size (number of pigs), duration, and spatial catchment: (i) local jackpot shows (n = 4), which tend to be relatively small (<300 pigs) and short in duration (<2 days) and draw exhibitors from primarily the host or neighboring states; (ii) national/regional jackpot shows (n = 6), which can attract thousands of pigs from across the United States; (iii) state fairs (n = 5), which often last 7 to 10 days; and (iv) county fairs (n = 98), which are limited only to pigs from that county but often commingle swine for an entire week. During sampling at the more geographically diverse jackpot shows, U.S. postal codes were collected with each sample corresponding to the location of the home farm for that exhibitor. Zip codes allowed us to investigate the geographic dispersal of IAVs across the country while maintaining the anonymity of swine exhibitors.
Snout wipe or nasal swab samples were collected from swine at each exhibition, placed in viral transport medium, and kept frozen at −80°C until testing as previously described (20, 29). Samples were collected from swine under The Ohio State University Institutional Animal Care and Use Committee approved protocol number 2009A0134-R2. Viral RNA was extracted from the samples and screened by real-time reverse transcriptase PCR (RT-PCR) using the VetMAX‐Gold SIV detection kit (Life Technologies) with the 7500 Fast real‐time PCR system (Life Technologies), following the manufacturer's protocol. All samples that were positive for IAV by RT-PCR were subsequently inoculated onto MDCK cells for virus isolation as previously described (30). Whole-genome sequencing was completed in the field and laboratory as previously described (14, 31) or by the Distributed Influenza Genomic Sequencing (DIGS) program (32). All genetic sequences were submitted to GenBank via the Influenza Research Database (33).
Phylogenetic analysis of IAVs.
To obtain a more complete picture of the IAV transmission between commercial swine, exhibition swine, and humans, we downloaded multiple sets of background genetic sequence data from publicly available databases. All available whole-genome sequence data for IAVs collected from swine in the United States during 2018 were downloaded from the National Center for Biotechnology Information’s Influenza Virus Resource, which curates data submitted to GenBank (34). The vast majority of IAV sequences submitted from swine in the United States to GenBank are from commercial swine, but we investigated a dozen sequences that we suspected were from exhibition swine, based on the time and location of virus collection, as well as genetic relatedness to viruses obtained from exhibition swine on the phylogenetic tree. We identified a small number of viruses that were likely to be from exhibition swine (T. Anderson, personal communications). In addition, we included IAV sequences collected in humans in the United States that are available from the Global Initiative on Sharing All Influenza Data’s (GISAID) EpiFlu database. Of the 1,720 H3N2 and 436 H1N1/H1N2 seasonal human viruses that were collected during summer months (defined as June 1 to September 30) during the years 2010 to 2018, 210 viruses were identified that were genetically related to U.S. swine viruses, including 10 from 2018 (nine H1N2 viruses and one H3N2 virus). Additional data on the larger number of lab-confirmed variant cases for which genetic sequence data were not available were obtained at the CDC’s interactive FluView site (https://gis.cdc.gov/grasp/fluview/Novel_Influenza.html).
Sequence alignments were generated separately for each genome segment (and separately for H1, N1, H3, and N2) and aligned using MUSCLE v3.8.3 (35), with manual correction in Se-Al v2.0 (available at http://tree.bio.ed.ac.uk/software/seal/). Phylogenetic relationships were inferred using the maximum-likelihood (ML) method available in the program RAxML v7.2.6 (36), incorporating a general time-reversible (GTR) model of nucleotide substitution with a gamma-distributed rate variation among sites. Due to the size of the data set, we used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (https://hpc.nih.gov/). To assess the robustness of each node, a bootstrap resampling process was performed (500 replicates), again using the ML method available in RAxML and the Biowulf Linux cluster. Separate viral introductions were identified on the phylogeny using clades defined by bootstrap values of >70. Maps were generated in the TileMill program (version 10.1; https://www.mapbox.com/tilemill/). Lineage assignment was performed using reference sequences as well as the H1 swine classification tool (37), available at the Influenza Research Database (https://www.fludb.org/) (33), and octoFlu (38), available at GitHub (https://github.com/flu-crew/octoflu).
Bayesian analysis of H1δ-2 viruses.
To further investigate the evolutionary relationships of H1δ-2 viruses in U.S. commercial swine prior to their detection in exhibition swine and humans in 2018, an additional data set was downloaded from the Influenza Virus Resource (34) that included all H1 sequences available for U.S. swine from the year 2005, when H1δ-2 viruses were first identified in swine (18), to 2019. Of the 6,699 H1 sequences available from U.S. swine during 2005 to 2019, 803 were identified in commercial swine that belong to the H1δ-2 lineage. The 803 sequences from GenBank were combined with 147 H1δ-2 sequences collected from U.S. exhibition swine during 2013 to 2018 and were used in subsequent analyses, using methods of alignment described above. TempEst v1.5.3 (39) was used to identify potential sequencing errors that substantially deviated from the linear regression of root-to-tip genetic distance against time. We performed a time-scaled Bayesian analysis using the Markov chain Monte Carlo (MCMC) method available via the BEAST v1.10.4 package (40) using the Biowulf Linux cluster. A relaxed uncorrelated lognormal (UCLN) molecular clock was used with a constant demographic, and a general time-reversible (GTR) model of nucleotide substitution with gamma-distributed rate variation among sites. The MCMC chain was run separately three times for each of the data sets for at least 300 million iterations with subsampling every 30,000 iterations using the BEAGLE 3 library to improve computational performance (41). All parameters reached convergence, as assessed visually using Tracer v.1.7.1, with statistical uncertainty reflected in values of the 95% highest posterior density (HPD). At least 10% of the chain was removed as burn-in, and runs for the same segment were combined using LogCombiner v1.10.4 and downsampled to generate a final posterior distribution of 1,000 trees that was used in subsequent analyses.
To infer and visualize viral gene flow between locations and types of swine population, a phylogeographic approach was used (42). The location state was specified for each viral sequence, allowing the expected number of location state transitions in the ancestral history conditional on the data observed at the tree tips to be estimated using “Markov jump” counts (43), which provided a quantitative measure of asymmetry in gene flow between defined populations. Viruses were categorized into three groups: (i) commercial swine from North Carolina, where the H1δ-2 virus originated (15); (ii) commercial swine from the Midwest and other U.S. regions (excluding North Carolina); and (iii) exhibition swine in all U.S. regions. For computational efficiency, the phylogeographic analysis was run using an empirical distribution of 1,000 trees, allowing the MCMC chain to be run for 25 million iterations, sampling every 1,000. A Bayesian stochastic search variable selection (BSSVS) was employed to improve statistical efficiency, since an asymmetric substitution model was used. Maximum clade credibility (MCC) trees were summarized using TreeAnnotator v1.10.4 and visualized in FigTree v1.4.4. To estimate the minimum distance traveled by H1δ-2 viruses in exhibition swine in 2018, we used the traveling salesperson problem (TSP) to determine the shortest tour that visits the 38 agricultural fairs at which H1δ-2 viruses were detected (TSP package in R) (44). Among all nearest-neighbor, insertion, and tour improvement heuristics available in the TSP package, the “cheapest-insertion” algorithm produced the shortest route between all fairs. The cheapest-insertion algorithm chooses a location (k) in each step such that the cost of inserting the new location (i.e., the increase in the tour's length) is minimal.
Data availability.
All sequences generated as a part of this study have been deposited in NCBI GenBank. Accession numbers are available in Table S1.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the agricultural fairs and swine shows that participate in our surveillance program as well as lab staff for their technical expertise and assistance in collection and testing of surveillance samples.
This work was supported by Centers of Excellence for Influenza Research and Surveillance, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contracts HHSN272201400006C and HHSN272201400008C.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. doi: 10.1128/MMBR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Kash JC, Morens DM. 2019. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci Transl Med 11:eaau5485. doi: 10.1126/scitranslmed.aau5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peiris JSM, Cowling BJ, Wu JT, Feng L, Guan Y, Yu H, Leung GM. 2016. Interventions to reduce zoonotic and pandemic risks from avian influenza in Asia. Lancet Infect Dis 16:252–258. doi: 10.1016/S1473-3099(15)00502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernández R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovão NS, Lozano-Dubernard B, Rambaut A, Van Bakel H, García-Sastre A. 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5:e16777. doi: 10.7554/eLife.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luis AD, Hayman DTS, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JRC, Mills JN, Timonin ME, Willis CKR, Cunningham AA, Fooks AR, Rupprecht CE, Wood JLN, Webb CT. 2013. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci 280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7(Suppl 4):42–51. doi: 10.1111/irv.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh B, Scheftel J, Bender J, Blackmore C, Bowman AS, Burkgren T, Calico M, Ellis D, Finelli L, Forshey T, Garvey A, Kasari E, Koeman J, Lauxman L, Marchall D, Paul M, Trock S, Uyeki TM. 2014. Measures to minimize influenza transmission at swine exhibitions. National Assembly of State Animal Health Officials and National Association of State Public Health Veterinarians.
- 8.Bowman AS, Nelson SW, Page SL, Nolting JM, Killian ML, Sreevatsan S, Slemons RD. 2014. Swine-to-human transmission of influenza A(H3N2) virus at agricultural fairs, Ohio, USA, 2012. Emerg Infect Dis 20:1472–1480. doi: 10.3201/eid2009.131082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson MI, Wentworth DE, Das SR, Sreevatsan S, Killian ML, Nolting JM, Slemons RD, Bowman AS. 2016. Evolutionary dynamics of influenza A viruses in US exhibition swine. J Infect Dis 213:173–182. doi: 10.1093/infdis/jiv399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanton L, Wentworth DE, Alabi N, Azziz-Baumgartner E, Barnes J, Brammer L, Burns E, Davis CT, Dugan VG, Fry AM, Garten R, Grohskopf LA, Gubareva L, Kniss K, Lindstrom S, Mustaquim D, Olsen SJ, Roguski K, Taylor C, Trock S, Xu X, Katz J, Jernigan D. 2017. Update: influenza activity—United States and worldwide, May 21–September 23, 2017. MMWR Morb Mortal Wkly Rep 66:1043–1051. doi: 10.15585/mmwr.mm6639a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, Boulton R, Haddy L, Biggerstaff M, Brammer L, Trock S, Burns E, Gomez T, Wong KK, Katz J, Lindstrom S, Klimov A, Bresee JS, Jernigan DB, Cox N, Finelli L, Influenza A (H3N2)v Virus Investigation Team . 2013. Human infections with influenza A(H3N2) variant virus in the United States, 2011–2012. Clin Infect Dis 57:S4–S11. doi: 10.1093/cid/cit272. [DOI] [PubMed] [Google Scholar]
- 12.Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, Beaudoin A, Berman L, Bidol S, Blanton L, Blythe D, Brammer L, D'Mello T, Danila R, Davis W, de Fijter S, Diorio M, Durand LO, Emery S, Fowler B, Garten R, Grant Y, Greenbaum A, Gubareva L, Havers F, Haupt T, House J, Ibrahim S, Jiang V, Jain S, Jernigan D, Kazmierczak J, Klimov A, Lindstrom S, Longenberger A, Lucas P, Lynfield R, McMorrow M, Moll M, Morin C, Ostroff S, Page SL, Park SY, Peters S, Quinn C, Reed C, Richards S, Scheftel J, Simwale O, Shu B, et al. 2013. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis 57:1703–1712. doi: 10.1093/cid/cit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow EJ, Davis CT, Elal AIA, Alabi N, Azziz-Baumgartner E, Barnes J, Blanton L, Brammer L, Budd AP, Burns E, Davis WW, Dugan VG, Fry AM, Garten R, Grohskopf LA, Gubareva L, Jang Y, Jones J, Kniss K, Lindstrom S, Mustaquim D, Porter R, Rolfes M, Sessions W, Taylor C, Wentworth DE, Xu X, Zanders N, Katz J, Jernigan D. 2018. Update: influenza activity — United States and worldwide, May 20–October 13, 2018. MMWR Morb Mortal Wkly Rep 67:1178–1185. doi: 10.15585/mmwr.mm6742a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambo-Martin BL, Keller MW, Wilson MM, Nolting JM, Anderson TK, Vincent AL, Bagal UR, Jang Y, Neuhaus EB, Davis CT, Bowman AS, Wentworth DE, Barnes JR. 2020. Influenza A virus field surveillance at a swine-human interface. mSphere 5:e00822-19. doi: 10.1128/mSphere.00822-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson MI, Lemey P, Tan Y, Vincent A, Lam TT-Y, Detmer S, Viboud C, Suchard MA, Rambaut A, Holmes EC, Gramer M. 2011. Spatial dynamics of human-origin H1 influenza a virus in North American swine. PLoS Pathog 7:e1002077. doi: 10.1371/journal.ppat.1002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bliss N, Stull JW, Moeller SJ, Rajala-Schultz PJ, Bowman AS. 2017. Movement patterns of exhibition swine and associations of influenza A virus infection with swine management practices. J Am Vet Med Assoc 251:706–713. doi: 10.2460/javma.251.6.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolting JM, Scheer SD, Bowman AS. 2019. Perceptions and attitudes of swine exhibitors towards recommendations for reducing zoonotic transmission of influenza A viruses. Zoonoses Public Health 66:401–405. doi: 10.1111/zph.12574. [DOI] [PubMed] [Google Scholar]
- 18.Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA, Janke BH. 2009. Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39:176–185. doi: 10.1007/s11262-009-0386-6. [DOI] [PubMed] [Google Scholar]
- 19.Stewart RJ, Rossow J, Conover JT, Lobelo EE, Eckel S, Signs K, Stobierski MG, Trock SC, Fry AM, Olsen SJ, Biggerstaff M. 2018. Do animal exhibitors support and follow recommendations to prevent transmission of variant influenza at agricultural fairs? A survey of animal exhibitor households after a variant influenza virus outbreak in Michigan. Zoonoses Public Health 65:195–201. doi: 10.1111/zph.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman AS, Workman JD, Nolting JM, Nelson SW, Slemons RD. 2014. Exploration of risk factors contributing to the presence of influenza A virus in swine at agricultural fairs. Emerg Microbes Infect 3:e5. doi: 10.1038/emi.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman AS, Nolting JM, Nelson SW, Slemons RD. 2012. Subclinical influenza virus A infections in pigs exhibited at agricultural fairs, Ohio, USA, 2009–2011. Emerg Infect Dis 18:1945–1950. doi: 10.3201/eid1812.121116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bliss N, Nelson SW, Nolting JM, Bowman AS. 2016. Prevalence of influenza A virus in exhibition swine during arrival at agricultural fairs. Zoonoses Public Health 63:477–485. doi: 10.1111/zph.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandbulte M, Spickler A, Zaabel P, Roth J. 2015. Optimal use of vaccines for control of influenza A virus in swine. Vaccines (Basel) 3:22–73. doi: 10.3390/vaccines3010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, Gauger PC, Loving CL, Webby RJ, García-Sastre A. 2012. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol 86:10597–10605. doi: 10.1128/JVI.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson MI, Culhane MR, Trovão NS, Patnayak DP, Halpin RA, Lin X, Shilts MH, Das SR, Detmer SE. 2017. The emergence and evolution of influenza A (H1α) viruses in swine in Canada and the United States. J Gen Virol 98:2663–2675. doi: 10.1099/jgv.0.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajão DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 89:11213–11222. doi: 10.1128/JVI.01675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walia RR, Anderson TK, Vincent AL. 2019. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 13:262–273. doi: 10.1111/irv.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan BS, DeBeauchamp J, Stigger-Rosser E, Franks J, Crumpton JC, Turner J, Darnell D, Jeevan T, Kayali G, Harding A, Webby RJ, Lowe JF. 2015. Influenza virus surveillance in coordinated swine production systems, United States. Emerg Infect Dis 21:1834–1836. doi: 10.3201/eid2110.140633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards JL, Nelson SW, Workman JD, Slemons RD, Szablewski CM, Nolting JM, Bowman AS. 2014. Utility of snout wipe samples for influenza A virus surveillance in exhibition swine populations. Influenza Other Respir Viruses 8:574–579. doi: 10.1111/irv.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowman AS, Nelson SW, Edwards JL, Hofer CC, Nolting JM, Davis IC, Slemons RD. 2013. Comparative effectiveness of isolation techniques for contemporary influenza A virus strains circulating in exhibition swine. J Vet Diagn Invest 25:82–90. doi: 10.1177/1040638712470449. [DOI] [PubMed] [Google Scholar]
- 31.Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. 2016. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 17:708. doi: 10.1186/s12864-016-3030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatesh D, Poen MJ, Bestebroer TM, Scheuer RD, Vuong O, Chkhaidze M, Machablishvili A, Mamuchadze J, Ninua L, Fedorova NB, Halpin RA, Lin X, Ransier A, Stockwell TB, Wentworth DE, Kriti D, Dutta J, van Bakel H, Puranik A, Slomka MJ, Essen S, Brown IH, Fouchier RAM, Lewis NS. 2018. Avian influenza viruses in wild birds: virus evolution in a multihost ecosystem. J Virol 92:e00433-18. doi: 10.1128/JVI.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Aevermann BD, Anderson TK, Burke DF, Dauphin G, Gu Z, He S, Kumar S, Larsen CN, Lee AJ, Li X, Macken C, Mahaffey C, Pickett BE, Reardon B, Smith T, Stewart L, Suloway C, Sun G, Tong L, Vincent AL, Walters B, Zaremba S, Zhao H, Zhou L, Zmasek C, Klem EB, Scheuermann RH. 2017. Influenza Research Database: an integrated bioinformatics resource for influenza virus research. Nucleic Acids Res 45:D466–D474. doi: 10.1093/nar/gkw857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 37.Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1:e00275-16. doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang J, Anderson TK, Zeller MA, Gauger PC, Vincent AL. 2019. octoFLU: automated classification for the evolutionary origin of influenza A virus gene sequences detected in U.S. swine. Microbiol Resour Announc 8:e00673-19. doi: 10.1128/MRA.00673-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayres DL, Cummings MP, Baele G, Darling AE, Lewis PO, Swofford DL, Huelsenbeck JP, Lemey P, Rambaut A, Suchard MA. 2019. BEAGLE 3: improved performance, scaling, and usability for a high-performance computing library for statistical phylogenetics. Syst Biol 68:1052–1061. doi: 10.1093/sysbio/syz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minin VN, Suchard MA. 2008. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol 56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 44.Hahsler M, Hornik K. 2007. TSP—infrastructure for the traveling salesperson problem. J Stat Softw 23:1–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated as a part of this study have been deposited in NCBI GenBank. Accession numbers are available in Table S1.