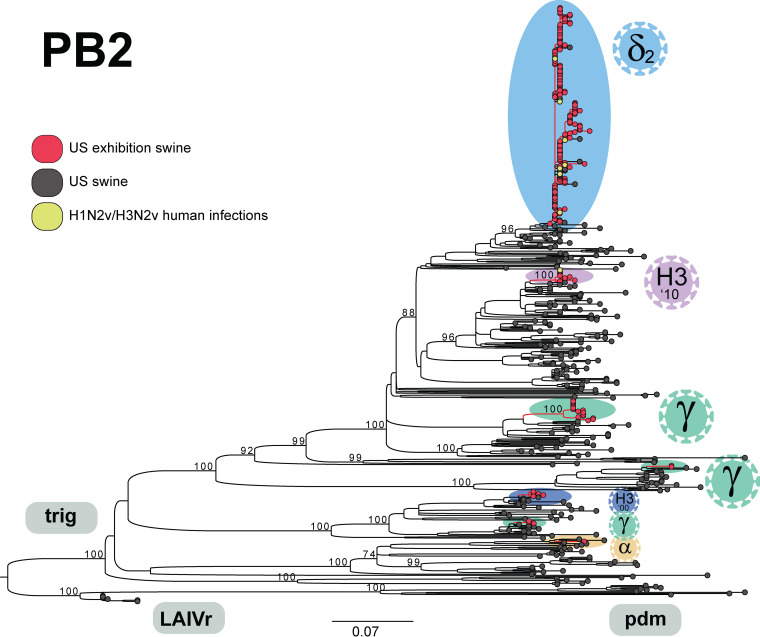
FIG 5.
Phylogenetic relationships of IAVs collected in U.S. swine during 2018. Maximum-likelihood tree inferred for PB2 segments (n = 526) of IAVs collected in the United States during 2018. Viruses collected for this study from U.S. exhibition swine are shaded red (n = 212); background sequences downloaded from GenBank are primarily from U.S. commercial swine and shaded black (n = 304); human infections with variant swine-origin viruses (H1N2v or H3N2v) were downloaded from GISAID and are shaded yellow (n = 10). All horizontal branch lengths are drawn to scale (nucleotide substitutions per site). The tree is midpoint rooted for clarity, and bootstrap values of >70 are provided for key nodes. Seven discrete introductions of IAVs into exhibition swine during 2018 are labeled by HA lineage (shading similar to that in Fig. 1).