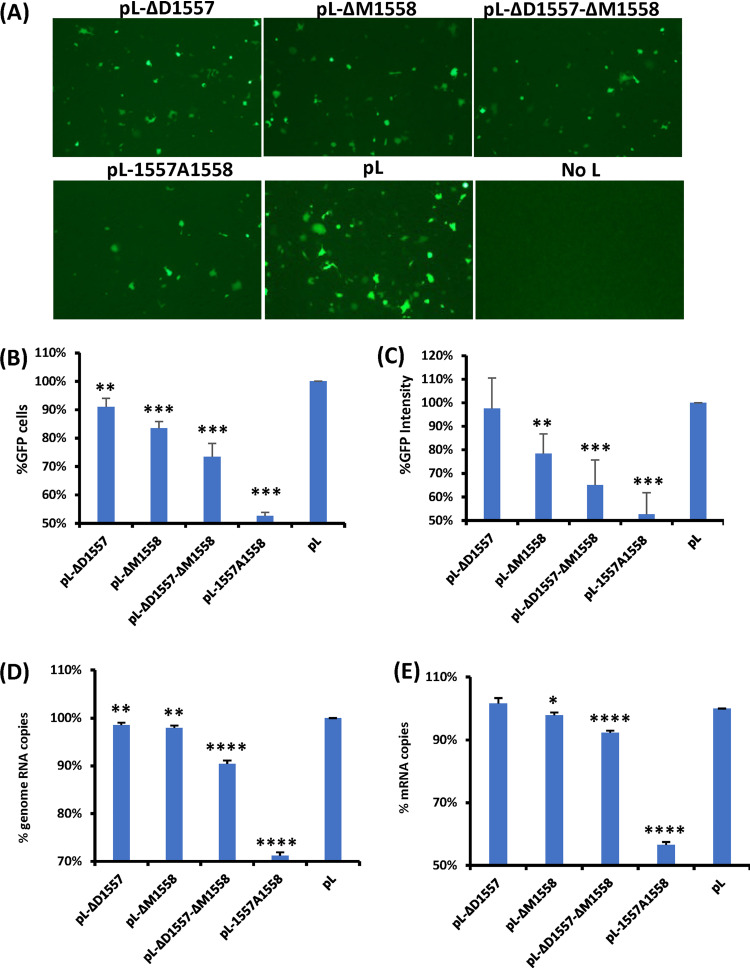
FIG 2.
Examination of the function of L deletion and insertion mutants using a minigenome assay. (A) L deletion and insertion mutants diminished GFP expression. Confluent HEp-2 cells were transfected with the minigenome plasmid together with pTM1-N, pTM1-P, pTM1-M2.1, pTM1-L, or a pTM1-L mutant, using Lipofectamine 2000. GFP expression was visualized by fluorescence microscopy at 48 h posttransfection. (B and C) Quantification of GFP-positive cells (B) and GFP intensity (C) by flow cytometry. HEp-2 cells were transfected with the minigenome and support plasmids. At 48 h posttransfection, cells were trypsinized, and the percentages of GFP-positive cells (B) and GFP intensity (C) were counted by flow cytometry. (D and E) Quantification of genome (D) and mRNA (E) of minigenome assay by real-time RT-PCR. HEp-2 cells were transfected with the minigenome and support plasmids. At 48 h posttransfection, cells were lysed by TRIzol reagent, and total RNA was extracted. Purified RNA was further treated by DNase to remove potential contamination of plasmid DNA. Then the genome (D) and mRNA (E) of the minigenome were quantified by real-time RT-PCR. Data for each L mutant were normalized by wild-type pL. The average from three independent experiments ± standard deviation is shown. NS, no significant difference (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.