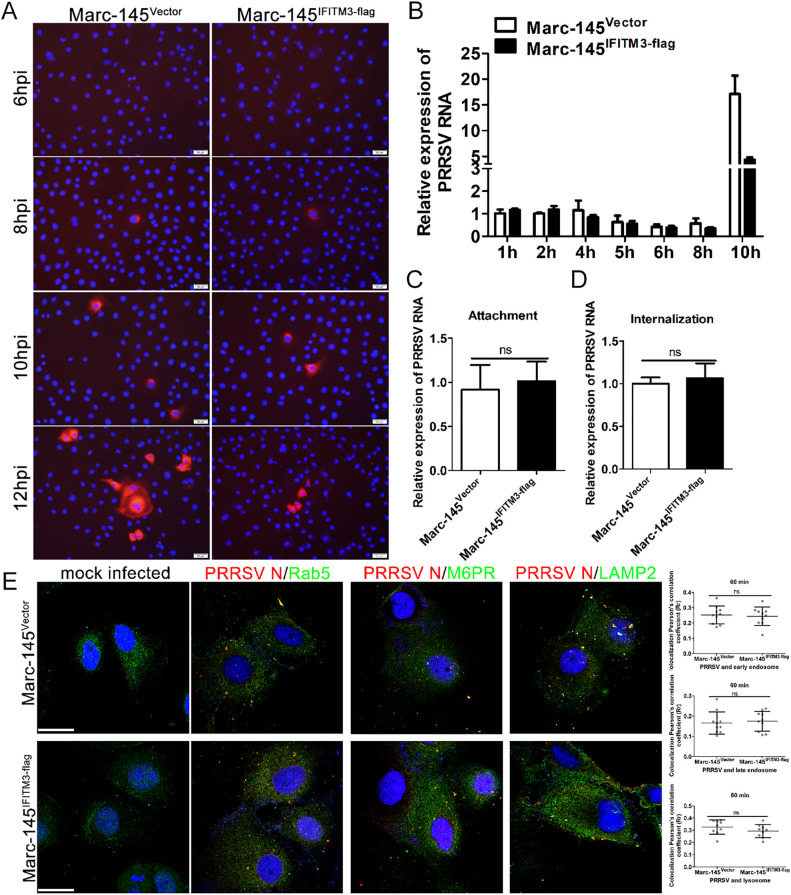
FIG 3.
IFITM3 does not affect PRRSV particle attachment, entry, or access to endosomes or lysosomes. (A) Marc-145-Vector or Marc-145-IFITM3-flag cells were incubated with PRRSV at an MOI of 1.0 for 6, 8, 10, and 12 h. The infected cells were fixed and stained with anti-PRRSV N antibody and counterstained with DAPI to visualize the nuclei. (B) Marc-145-Vector or Marc-145-IFITM3-flag cells were infected with 1.0 MOI of PRRSV for 1 h at 37°C, and then cells were washed with PBS and cultured with 3% FBS+DMEM. Cells were harvested 1, 2, 4, 5, 6, 8, or 10 hpi, and PRRSV genome levels were determined. Prechilled Marc-145-Vector or Marc-145-IFITM3-flag cells were infected with PRRSV (1.0 MOI) and further chilled on ice for 1 h. (C) For the attachment assay, after washing 3 times using ice-cold PBS, cells were harvested for PRRSV genome abundance analysis. (D) For the entry assay, cells were washed with PBS three times and treated with 37°C prewarmed DMEM and incubated at 37°C for 1 h, followed by trypsin treatment for 30 sec and 3 washes with PBS. Cells were collected for PRRSV genome content analysis. (E) Marc-145-Vector or Marc-145-IFITM3-flag cells were inoculated with 1.0 MOI PRRSV for 1 h at 37°C, and then cells were fixed and stained with anti-PRRSV N, -Rab5, -M6PR, and -LAMP2 antibodies, followed by an Alexa Fluor 488-conjugated goat anti-rabbit IgG (H&L) antibody and an Alexa Fluor 594-conjugated goat anti-mouse IgG (H&L) antibody. Colocalization of PRRSV particles with cellular vesicles was quantified, and representative images are presented. Scale bar, 10 μm. Horizontal bars represent the mean of the Pearson’s correlation coefficient (Rr) calculated based on 10 fields of view, with error bars marking the 95% confidence intervals. PRRSV, porcine reproductive and respiratory syndrome virus; MOI, multiplicity of infection; hpi, hours postinfection.