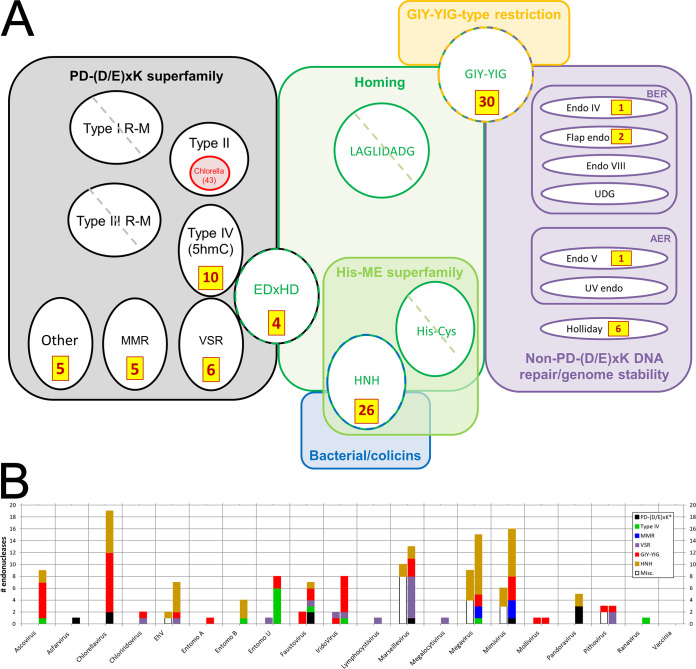
FIG 5.
Structure-based deep mining markedly elevates numbers of endonucleases identified across the NCLDV. All accession numbers are given in Table S3 in the supplemental material. (a) Counts of newly identified NCLDV endonucleases (brown/yellow boxes) mapped onto known DNA endonuclease structural/functional classes (colored boxes/circles/ovals). Circles/ovals with no counts shown indicate major classes with no representatives reported among any NCLDV (diagonal hatch) or none newly identified here (no hatch). PD-(D/E)xK (black), structural superfamily showing the following functional classes: REases (types I to IV), nicking endonucleases for DNA mismatch repair (MMR), and very short patch repair (VSR), and one class of homing endonucleases (EDxHD). The VSR, type IV, and EDxHD groups are shown touching to illustrate their particularly close structural relationship in search results (see the text). Types I and III REase polypeptides are denoted “R-M” due to their dual function as restriction-modifying enzymes, in which we focused only on the “catalytic site for DNA cleavage” (96) and “endonuclease domain” (97), respectively. Red indicates strain depth dimensionality for Chlorella virus type II REases specifically (from REBASE rather than UniProt; see text). Other major colors indicate functional classes encompassing either structural families (green, blue, and orange) or functional classes (purple). In the latter, BER and AER represent base and alternative excision repair, respectively; UDG, uracil DNA glycosylase (an endonuclease); Endo IV, AP-endonuclease. Counts do not include our assignments/reassignments of previously identified/annotated endonucleases (see the text). (b) NCLDV endonuclease counts by virus. Entomopox A, B, and U refer to entomopox alpha, beta, and unclassified, respectively. Bars to the left and right of the central tick represent counts before and after deep mining, respectively. “Before” counts include proteins that matched an endonuclease here and were also “endonuclease” or “nuclease” according to UniProt gene_name. “After” counts represent “before” counts plus endonucleases newly identified here plus reassignments (see the text). Each bar is divided by color according to endonuclease class (see color legend). The PD-(D/E)xK* class refers to PD-(D/E)xK homing plus “Other”” (panel a). The “Misc.” class (“before”) contains, exclusively, members reassigned to other classes in the “after” sections based on primary structural homolog (see the text). Excluded from the graph are all Chlorella virus “red ring” (panel a) restriction endonucleases not from PBCV-1 (Table 1). For simplicity, the small numbers of repair endonucleases (purple section of panel a) are omitted. Counts represent “top hit only” structural matches).