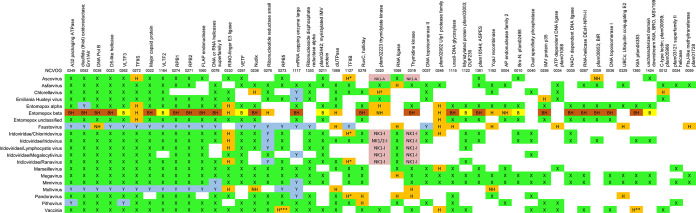
TABLE 2.
Expanded coverage by structure-based deep mining of 47 genes previously designated NCVOGsa
NCVOGs (4, 21, 22, 24), conserved among NCLDV on the basis of protein sequence homology, are ordered (left to right) by descending coverage among our 20 NCLDV. NCVOGs are named according to additional file 4 in Yutin et al. (24). “X” (green) indicates prior coverage on the basis of sequence homology (see references 21 and 25) and references therein); “Y” (lilac) indicates NCVOG additions in Koonin and Yutin (21). “H” (mustard) indicates new coverage via structure-based deep mining. H*, TFIIB structural homolog lacking zinc finger domain. H**, known KilA-N domain protein from literature (vaccinia protein p28 [67, 102, 103]). H***, known RPB5 homolog from literature (vaccinia protein RP22 [15]). “NH” indicates deep mining result supported by UniProt protein name (not present in prior NCVOG analyses). Since prior NCVOG analyses did not include entomopox beta, coverage for this virus was partially elucidated by BLASTP search (“B,” yellow). “BH” (brown) indicates combination of BLASTP and deep mining. NK1, NK2 (pink), nucleoside kinase-type (NK) proteins are dually listed between NCVOGs 0319 and 0320 to cover both the original designation and our interpretation (see Table S2 in the supplemental material). Coverage may appear low for some NCVOGs since they were designated as such on the basis of all known NCLDV (25) as opposed to the 20 representatives considered here.