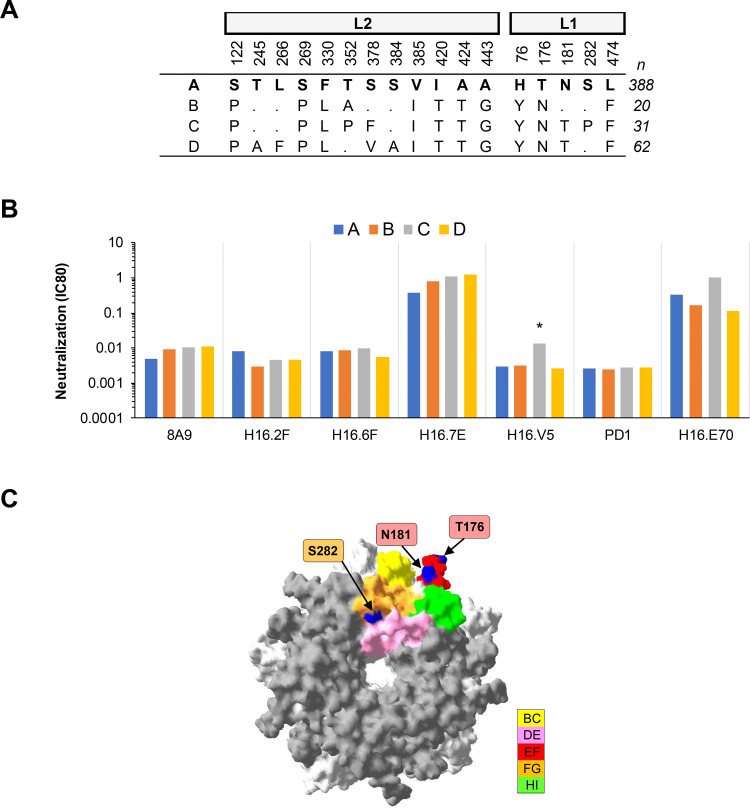
FIG 1.
Neutralization of HPV16 lineage variants by MAbs. (A) Consensus lineage HPV16 L1L2 variant sequences used for creation of representative PsVs (29). (B) Neutralization sensitivity of lineage L1 and L2 PsV variants to HPV16 MAbs. Only MAbs that had an IC80 of <0.5 μg/ml were included. The symbol * indicates a ≥4-fold reduced sensitivity in the magnitude of difference in MAb sensitivity (IC80) between the indicated variant and the consensus A/A1 PsV. (C) HPV16 lineage A homology model based upon the HPV16 L1 pentamer crystal (Protein Databank [PDB] accession number 2R5H.1; using DeepView Swiss-Pdb viewer v4.0) with top view shown. The quality of a predicted model was assessed by their qualitative model energy analysis (QMEAN4) and global model quality estimation (GMQ), which were −3.33 and 0.99, respectively. All five L1 monomers of the pentamer are pictured, with external loops (gray) and all visible sites of variation from the consensus A/A1 (blue) highlighted as indicated (yellow, BC1; pink, DE2; red, EF2; orange, FG2; and green, HI1).