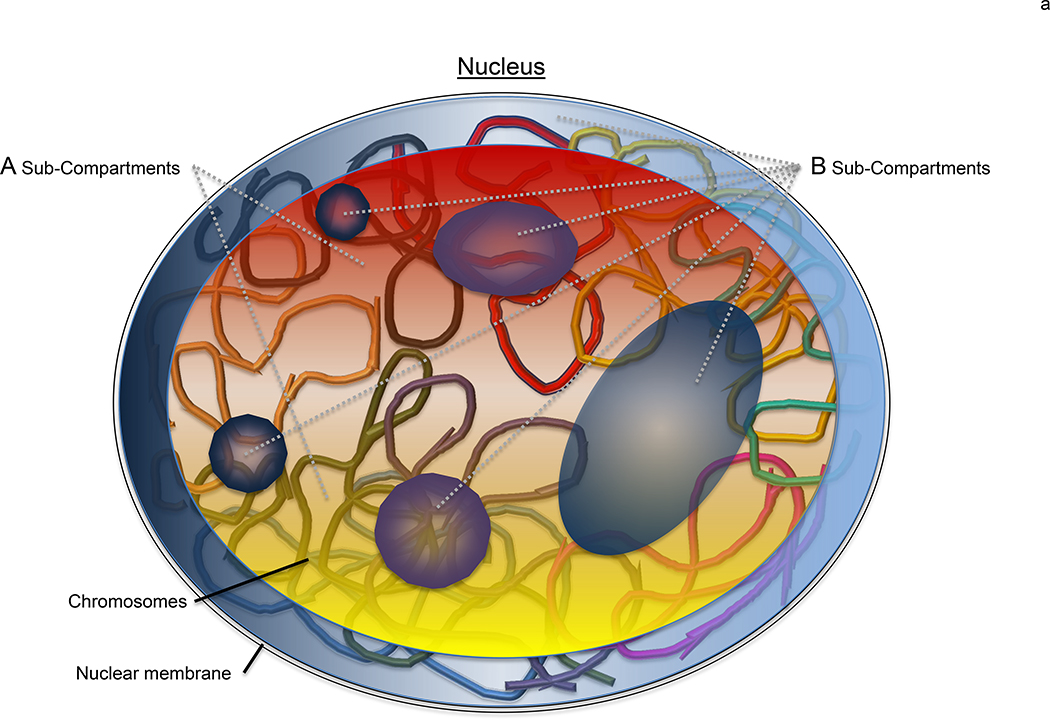
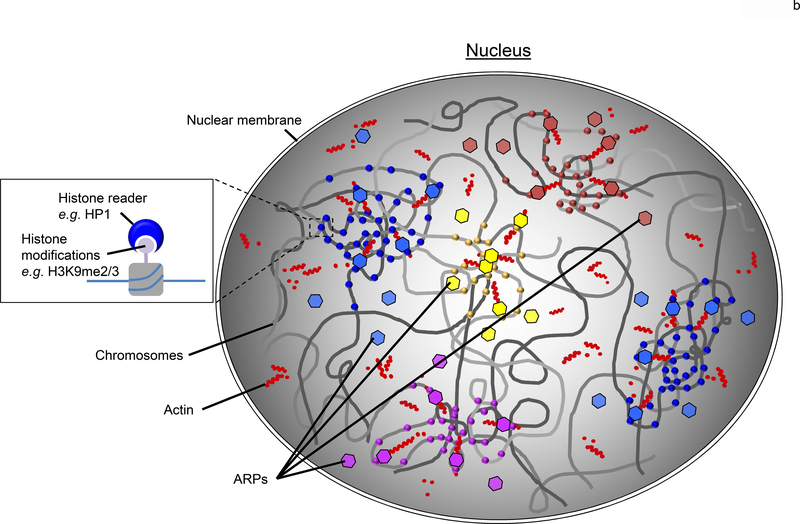
Figure 1. Chromosome regions are clustered based upon epigenetic signatures.
(a) In a nucleus, eukaryotic genomes are partitioned into two main compartments (A/B), as detected by Hi-C [6], that correspond to the classic heterochromatin and euchromatin regions [7]. The main compartments are further sub-divided (compartments A1-A2 and B1-B4) based upon varying epigenetic signatures. Here, the general locale of A1 and A2 sub-compartments are represented by the red and yellow regions primarily found in the nuclear interior whereas the B1-B4 sub-compartments are shown by the blue and purple areas that are minimally enriched at the nuclear periphery and around the nucleolus. While it is clear that the various regions correlate with select epigenetic modifications [11,16], how this information is used to physically cluster the chromatin is yet to be revealed. (b) A plausible model to account for the nuclear organization of large-scale genomic features such as the A/B compartments and sub-compartments (A1-B4) is the coordinated use of a dynamic nuclear actin network to effectively move the large chromosome regions, epigenetic readers (eg, HP1) to selectively distinguish the various histone marks (circles), and ARPs (hexagons) to bridge the readers to the actin (red dots) network. The various colors of the circles and hexagons, represent the various readers and ARPs found with eukaryotic nuclei. Together, this system would provide an effective means to reorganize a genome after each cell division within a biologically useful timescale (<hour).