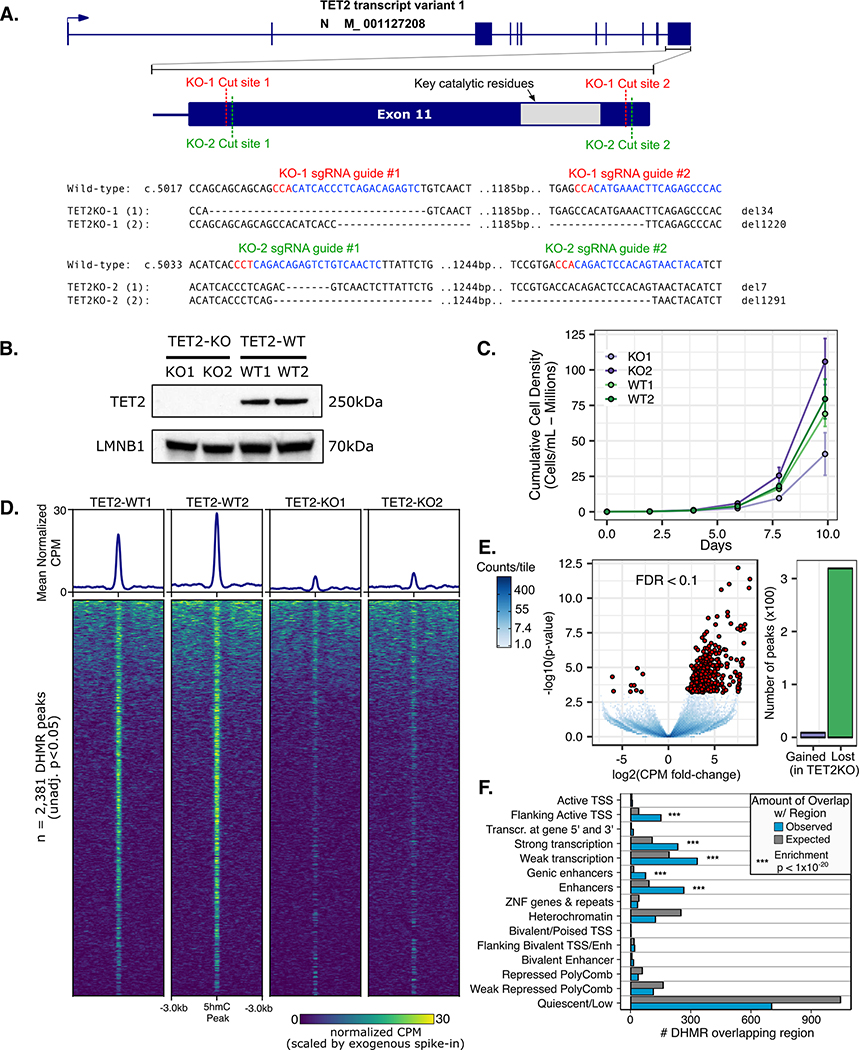
Figure 1. CRISPR/Cas9 mediated loss of TET2 alters growth and hydroxymethylation patterns in TF1.
A) Paired CRISPR sgRNA design targeted key catalytic residues of TET2. Two independently derived TF1 clones with bi-allelic inactivating deletions were characterized. B) Western blot using C-terminal anti-TET2 antibody confirms loss of full-length TET2 protein. C) Growth curves for TF1 clones display differences in normal growth patterns for TET2 knockout. D) 5-hmC pull down and sequencing (HMCP) peak heatmaps and mean normalized counts profile for differentially hydroxymethylated regions (DHMR). E) Differential 5hmC peak enrichment volcano plot TET2KO vs. WT. F) Number of DHMR (TET2KO v. WT) observed vs. expected overlapping regulatory chromatin states in reference CD34+ HSC epigenome (from reference epigenome E051 of Roadmap Epigenomics Consortium)