Fig. 1. Initial screen of CGBE candidates for C:G to G:C editing.
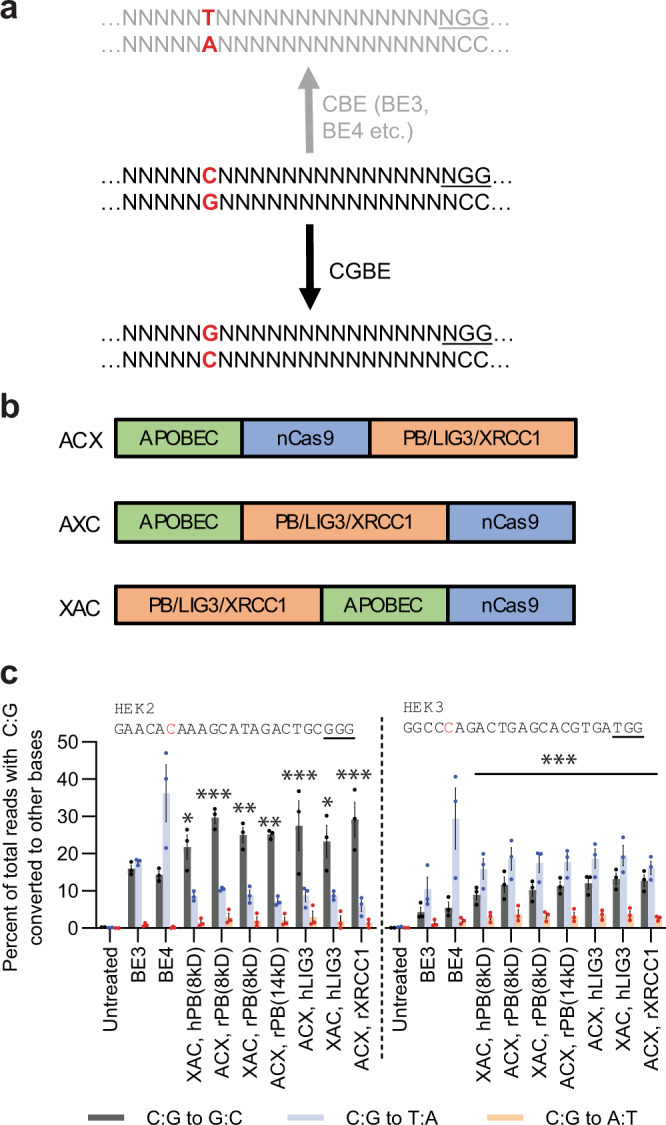
a CBEs like BE3 and BE4 predominantly convert C:G to T:A while CGBE aims to predominantly convert C:G to G:C. b CGBE candidates were designed in three orientations – ACX, AXC, and XAC, where X denotes the fused BER protein. c Seven candidates were selected for their high C:G to G:C editing at both HEK2 and HEK3. The lower editing at HEK3 is likely due to a disfavored motif (refer to data in Fig. 2a). Targeted C’s are in red. PAMs are underlined. *p < 0.05; **p < 0.01; ***p < 0.001 using one-way ANOVA (Dunn- Šidák) of C:G to G:C editing against ‘Untreated’. Exact p values are available in Source Data. Each dot represents editing of an individual biological replicate; bars represent mean values; error bars represent SEM of three biologically independent replicates.
