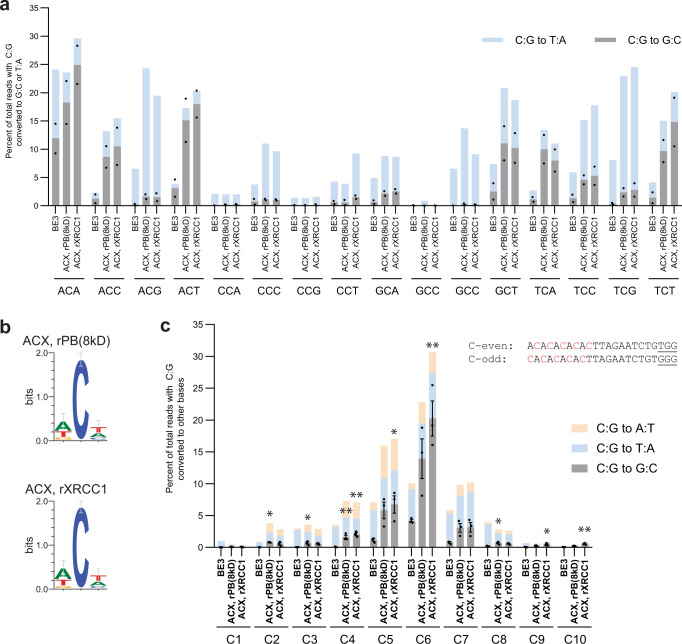
Fig. 2. Sequence context and editing window for two selected CGBEs.
a Evaluation of C:G to G:C editing at each NCN DNA motif. 16 different gRNAs were designed to target the genomic region around the HEK2 site (HEK2-1 to HEK2-16 in Supplementary Table 2), chosen such that the gRNA-to-target combinations together cover all NCN motif contexts and that genomic distance among gRNAs is minimized (14/16 gRNAs, including the initial HEK2-1 gRNA:target, reside within a 1.8 kb region, while the other 2 gRNAs target within 10 kb). Each dot represents C:G to G:C editing of an individual biological replicate; bars represent mean values of two biologically independent replicates. b DNA WebLogo created with target motifs in which C:G at position 6 was edited to G:C (n = 2 biologically independent replicates; error bars are Bayesian 95% confidence intervals). c Editing window of CGBEs using gRNAs with alternating 5′-W-C-3′ motifs. Targeted C’s are in red. PAMs are underlined. *p < 0.05; **p < 0.01 using one-way ANOVA (Dunn-Šidák) of C:G to G:C editing against ‘BE3’. Exact p values are available in Source Data. Each dot represents C:G to G:C editing of an individual biological replicate; bars represent mean values; error bars represent SEM of three biologically independent replicates.