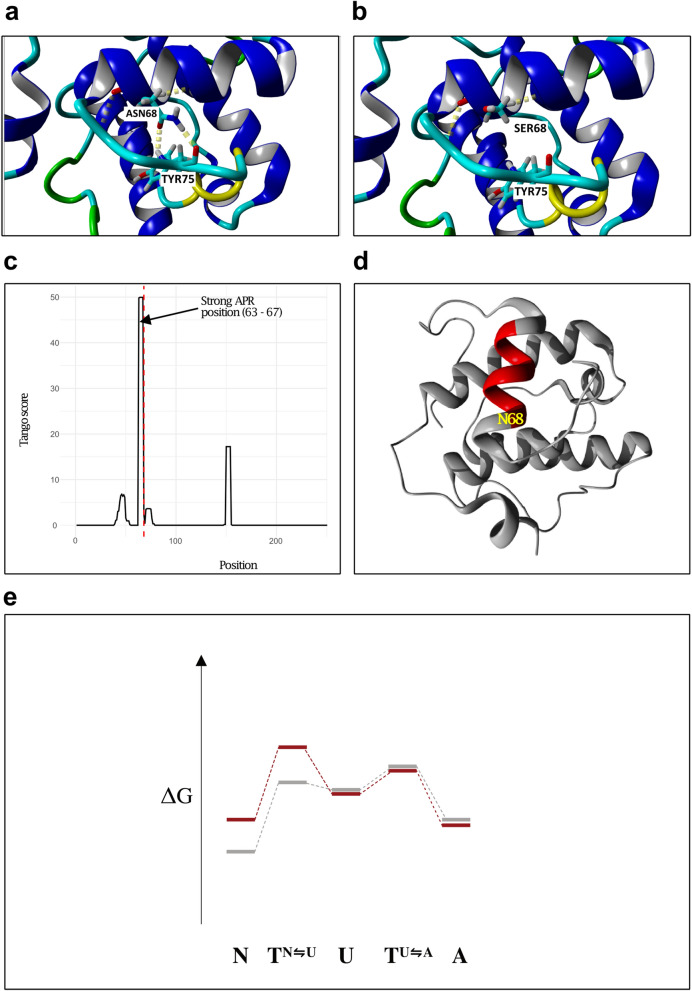
Figure 2.
Modelling of the MAPRE2 N68S mutation. (a, b) The size and hydrophobicity difference between (a) wild type (Asn) and (b) mutant (Ser) residue makes the mutant unable to form the same hydrogen bonds. (c) Aggregation propensity prediction TANGO scores per residue for wild type MAPRE2. Vertical red line indicates the position of N68. (d) Model structure of MAPRE2 with the strong APR colored in red. (e) Energetic landscape of the mechanism proposed. N68S mutation is indicated in red and wild type energy landscape in grey. N68S thermodynamically destabilizes and kinetically slows down the folding reaction, resulting in higher energies for N and TN⇋U and less natively folded protein (N native fold, U unfolded state, A aggregated state, T transition state).