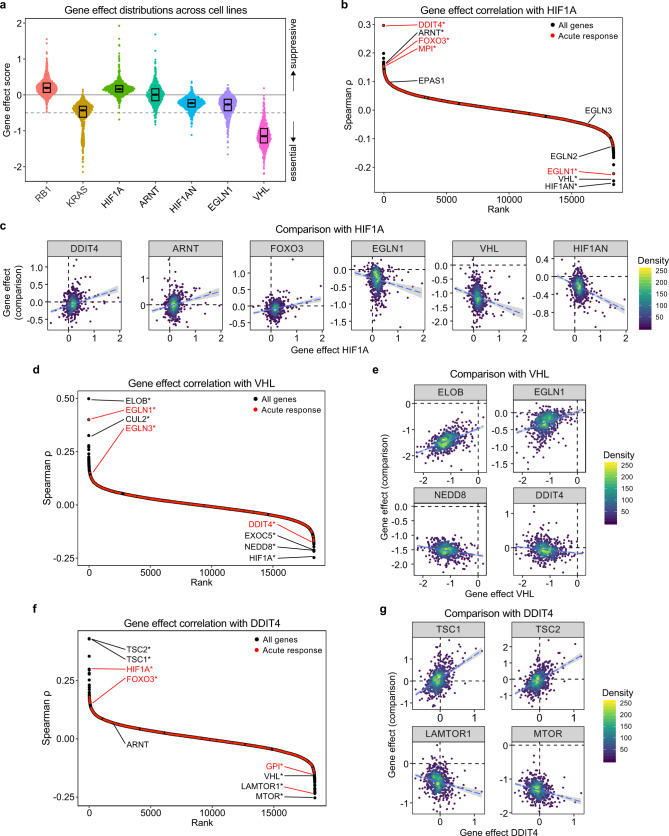
Fig. 6. HIF1A suppresses cancer cell viability in normoxia.
a Gene effect score distributions for HIF1A and select known regulators, alongside the tumor suppressor RB1 and oncogene KRAS for comparison, Horizontal spread of data points is proportional to density; boxes indicate medians and upper and lower quartiles; dashed line indicates threshold below which a gene is considered essential. b Ranked Spearman correlation coefficients of gene effect scores for each gene against HIF1A, with selected genes labeled, and acute response genes highlighted in red. * denotes significant correlations (FDR < 10%) — see Supplementary Data 7 for exact FDR values. c Comparison of gene effect scores with select genes for HIF1A. Points representing cell lines are colored by density and dashed lines represent linear fits to the data with 95% confidence intervals in gray. d Ranked Spearman correlation coefficients of gene effect scores for each gene against VHL, with selected genes labeled, and acute response genes highlighted in red. * denotes significant correlations (FDR < 10%) — see Supplementary Data 7 for exact FDR values. e Comparison of gene effect scores with select genes for VHL. Points representing cell lines are colored by density and dashed lines represent linear fits to the data with 95% confidence intervals in gray. f Ranked Spearman correlation coefficients of gene effect scores for each gene against DDIT4, with selected genes labeled, and acute response genes highlighted in red. * denotes significant correlations (FDR < 10%) — see Supplementary Data 7 for exact FDR values. g Comparison of gene effect scores with select genes for DDIT4. Points representing cell lines are colored by density and dashed lines represent linear fits to the data with 95% confidence intervals in gray. See also Supplementary Fig. 8 and Supplementary Data 7.