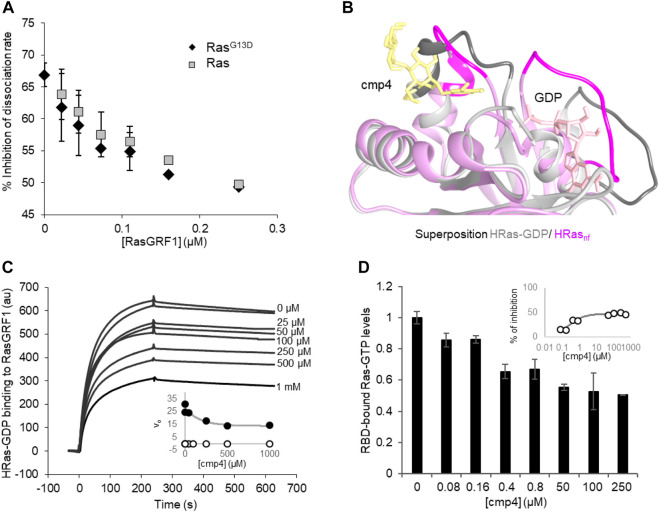
FIGURE 3.
cmp4 affects HRas binding to GEF (RasGRF1) and effector (Raf1-RBD) in a dose-dependent manner. (A) Inhibition of nucleotide dissociation rate on both HRas and HRasG13D (1 μM) in the presence of 100 μM cmp4 and increasing concentrations of RasGRF1 (range 0.01–0.25 μM). (B) Best fitting pose of cmp4 on HRas-GDP (pink) was superimposed to the structure of nucleotide-free Ras (HRasnf, in grey) from the crystal structure of the hSos1 catalytic domain associated with HRas (PDB ID: 1bkd). Switch I and II regions are stained darker. GDP is in pink, cmp4 in yellow; (C) Biacore-based direct measurement of 0.5 μM GEF (GST-RasGRF1) binding to His-HRas-GTP in the presence of increasing concentrations of cmp4 (25–1000 μM). In the insert kinetics analysis of RasGRF1 binding to HRas-GDP in the presence of different concentrations of cmp4, relative to SPR curves. All points for initial association rate (von, closed symbols, voff, open symbols) were fitted respectively to a nonlinear ‘growth-sigmoidal Hill’ curve (n = 1), which is reported in the graph as a thin line; (D) Levels of HRas-GTP bound to a Ras binding domain (RBD) of Raf1 in the presence of increasing concentrations of cmp4 (range 0–500 μM), detected with the G-LISA® kit (Cytoskeleton, Inc. BK131). Data were normalized to Ras-GTP levels measured in the absence of cmp4 (control). All data are significant at 99%, as calculated by Student’s t-test in comparison to control. In the inset, the percentage of inhibition of Ras-GTP bound to RBD as a function of cmp4 concentration, relative to the G-LISA experiment. All points were fitted respectively to a nonlinear ‘growth-sigmoidal Hill’ curve (n = 1).